Nowadays, the opportunistic pathogen,
Klebsiella pneumoniae (
K. pneumoniae) is considered as the main bacteria involved in nosocomial infections [1]. Infections caused by
K. pneumoniae including urinary tract infections, septicemia, pneumonia and intra-abdominal infections in hospitalized patients are responsible for a high rate of mortality [2]. Antibiotic resistance has always been regarded as a serious problem for human health by affecting patients in hospitals around the world. [3]. Considering that
K. pneumoniae causes disease in patients with a weakened immune system thereby increasing the rate of resistance to antibiotics in bacteria, it can be a serious threat for health care settings [2, 3]. Therefore, the determination of antibiotic resistance patterns in common pathogenic bacteria aimed at conducting empirical and specific therapy against a particular pathogen is important [4]. One of the most common antibiotic resistances in gram negative bacteria such as
K. pneumoniae is aminoglycoside resistance [5]. Antibiotics such as streptomycin, gentamicin, tobramycin, amikacin, and kanamycin are known as the family of amino- glycoside antibiotics. All aminoglycosides inhibit protein synthesis in bacteria by binding to the 30S subunit of 16S rRNA and thus show bactericidal effect [4, 5]. Production of aminoglycoside modifying enzymes (AMEs) is the most common type of resistance to aminoglycosides which results in a high level of bacterial resistance [6, 7]. The common encoding genes for aminoglycoside modifying enzymes in the Enterobacteriaceae family are
aac (3)- II and
aac (6')- Ib [8]. N-acetyltransferases
aac (6) and
aac (3) are most frequently found in clinical isolates in Iran and certain other countries [3,8-10]. Aminoglycoside 6′–N-acetyltransferases of type Ib [aac(6′)-Ib] are widespread among members of the Enterobacteriaceae family including
K. pneumoniae [11, 12]. There are limited studies on the AMEs in
K. pneumoniae in Iran, Considering the role of
K. pneumoniae in nosocomial infections as well as the importance of identifying resistance to aminoglycosides particularly resistances caused by mobile genetic elements, the aim of this study was to determine the two genes
acc (6')- Ib and
acc(3)-II in clinical isolates of
K. pneumoniae from patients admitted to hospitals in the city of Borujerd in the west of Iran.
Materials and Methods
K. pneumoniae isolates
In a cross-sectional study, a total of 100
K. pneumoniae strains were isolated from clinical samples (blood, wound, urine and trachea) of patients in the hospitals of Borujerd from April to September 2015.
K. pneumonia isolates were identified and confirmed by conventional microbiological tests: Gram staining and standard biochemical tests such as lactose fermentation, indole test, motility, citrate and urease test, lysine decarboxylase and
Methyl Red Voges Proskauer (MR-VP).
Antimicrobial susceptibility testing
The antimicrobial susceptibility of
K. pneumoniae
isolates to gentamicin (10 μg), amikacin (30 μg),
ampicillin (10 μg), cephalothin (30 μg)
aztreonam (30 μg)
ceftriaxone (30 μg),
chloramphenicol (30 μg), ciprofloxacin (5 μg), imipenem (10 μg), and nalidixic acid (30 μg) disks (Rosco company, Denmark) was determined by disk diffusion method, according to clinical and laboratory standards institute guidelines (9).
K. pneumoniae ATCC 13883 was used as the control strain for disk susceptibility testing.
Detection of aac(6')-Ib, aac(3)-II genes
Genomic DNA was extracted from all aminoglycoside resistance
K. pneumoniae isolates using a DNA extraction kit (Cinapure DNA, CinaClon, Iran) according to the manufacturer’s instructions. Amplification of the genes encoding aminoglcoside modifying enzymes,
aac(6')-Ib, and
aac(3)-II, was performed using specific primers (Table 1) by polymerase chain reaction (PCR). The PCR mixture was prepared in a final volume of 25 μl consisting of template DNA (1 μl), 0.25 μM of the respective primers [13], 2.5 μl PCR buffer, 0.5 μM deoxynucleotide triphosphates, 0.75 μM of MgCl2, 0.25 U Taq DNA polymerase (Cinna Gene, Tehran, Iran), and 19.5 dd H
2O. A thermocycler (PEQ STAR, Germany) was programmed with the following parameters: after an initial denaturation for 5 min. at 94°C, 30 cycles of amplification were performed with denaturation at 94°C for 30 sec, annealing at 55°C for 30 sec, and DNA extension at 72°C for 30 sec, followed by a final extension at 72°C for 5 min. Then, PCR products were visualized by electrophoresis on 1% agarose gel. This study was approved by Ethics Committee of Hamadan university of medical sciences, Hamadan, Iran.
Results
The results of this study showed a significant difference between the types of the clinical samples (urine) as regards resistant to aminoglycosides. A total of 79% (27 isolates) of aminoglycosides resistant isolates were isolated from urine and other clinical samples including trachea 11.7% (4 isolates), wounds 5.8% (2 isolates) and blood 2.9% (1 isolate). Based on the results of antibacterial sus- ceptibility testing, out of 100 samples of
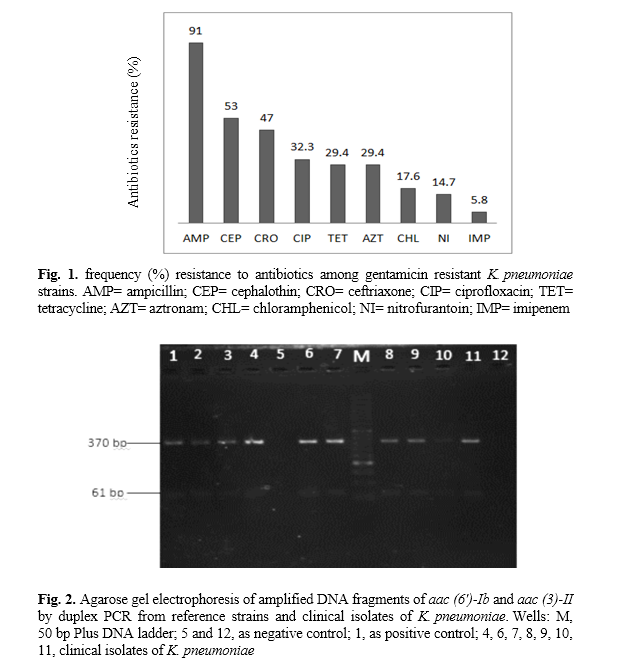
K. pneumoniae, 34% of the isolates were resistant to gentamicin and 21% to amikacin. Moreover, resistance to both gentamicin and amikacin was detected in 18% of the isolates. According to the results of the susceptibility tests, of the 34 gentamicin resistant isolates , 94% (32 isolates) were resistant to ampicillin and 51% to amikacin and ceftriaxone. The results of the antibiogram for the 34 gentamicin resisting isolates are shown in Figure 1.
The amplification of aminoglycoside resistant genes by PCR showed that 71% (n=24) and 5.8% (n=2) of the aminoglycoside resistant strains harbored the
aac (3)-IIa and
aac (6ʹ)-Ib genes; simultaneous harboring of the
aac (3)-IIa and
aac (
6ʹ) Ib genes was found in 64% (n=22) of the aminoglycoside resistant isolates (Fig. 2). In this study we could also detect the simultaneous presence of the
aac (6ʹ)- Ib and aac (3)- IIa genes in one PCR reaction (duplex PCR).
Emergence of resistant strains to aminoglycosides can be a serious threat against public health in hospitals and society, causing difficulties for medical treatment and imposing additional costs on the healthcare system. In this study, antibiotic resistance patterns and the occurrence frequency of the aminoglycoside resistance genes aac (6')-Ib and aac (3)-IIa in K. pneumoniae strains isolated from hospitalized patients in Borujerd, in the west of Iran, was investigated by PCR. These genes encode aminoglycoside modifying enzymes for gentamicin and amikacin. In fact, acetyltransferase enzymes of gentamicin and amikacin are encoded by these genes [10]. Different results concerning resistance to aminoglycosides have been published in Iran; however, few studies have been carried out on the prevalence of aminoglycoside resistant genes in K. pneumoniae [11, 12, 14, 15]. Linderman et al. from Norway reported that 90% of K. pneumoniae strains are resistant to aminoglycosides [16]. The results of our research indicated a relatively high resistance (34%) to gentamicin. Resistance to both gentamicin and amikacin was observed in 18% of the cases. Antibiotics that inhibit cell wall synthesis increase the transfer of aminoglycosides into bacteria cells, hence the combination of cell wall synthesis-inhibitor antibiotics such as beta-lactams and aminoglycosides can be used to treat infections caused by K. pneumonia [7, 12, 14]. In recent years, strains of bacteria resistant to beta-lactams and aminoglycoside antibiotics, especially in hospitals, have increased [17]. In this study, over 90% of the isolates resistant to gentamicin were also resistant to ampicillin. These results suggest that combining these two classes of antibiotics is not effective in treating resistant strains of K. pneumonia while the imipenem showed the highest activity against the most resistant strains to aminoglycosides. In contrast to our results, Peerayeh, et al. reported a higher prevalence (42.5%) of aac (6')- Ib genes among K. pneuominea isolates in Tehran’s hospitals. They also detected the aac (3)- II gene in 35.1% of these isolates [18]. In addition, Lindermann reported a higher frequency for the aac (3)-II gene found in 79.3% of K. pneumonia isolates in comparison with the aac (6')- Ib gene (found in 37.9% of the isolates) [16]. In 2015, Liang et al. studied aac (6')- Ib in K. pneumoniae isolates and found 19% of the isolates containing this gene [19].
In a study, Almaghrabi et al., reported resistance to gentamicin and amikacin in 40%, and 16% of K. pneumoniae isolates, respectively. In the United States' hospitals, 98% of amioglycoside resistant strains poss- essed AMEs, including aac (6ʹ)-Ib. These results have shown that the location of sampling may affect the distribution of amino- glycoside genes among K. pneomoniae isolates [20]. Other isolates with negative PCR results for AME genes have probably become resistant to aminoglycosides through other resistance mechanisms such as the reduction of uptake or change in the ribosome binding site [18, 21].
Conclusions
The results of this study indicate moderate resistance to aminoglycosides in comparison with the results achieved by other researchers. Furthermore, the presence of the aac (6')- Ib and aac (3)-IIa genes was observed in more than 50% of nosocomial K. pneumoniae strains resistant to aminoglycosides. This may be due to the transmission of this gene through mobile genetic elements such as plasmids and transposones that create a high risk of rapid spread of these genes among K. pneunmoniae isolates in hospitals.
Conflict of Interest
The authors declare no conflict of interest.
Acknowledgement
We would like to thank the staff of microbiology laboratories in the hospitals of Brojured, Iran.









































