Introduction
Chronic lymphocytic leukemia (CLL) is one of the most prevalent kinds of leukemia in the West, specified by mature appearing monoclonal B cells with CD5, CD19, and CD23 markers. B-lymphocytes mature in the bone marrow by rearrangement of the immunoglobulin variable gene which is applied as a B-cell antigen receptor [1]. The incidence of this type of leukemia is 30 percent among all blood cancer individuals and is slightly more common in men than in women. It has been reported that the leukemia causes ∼5,000 mortalities in the United States annually. One of the significant factors to be considered in the treatment of cancer, which helps to select better therapy options, is the early diagnosis. Detection of biomarkers at various levels such as transcriptomes and proteomics levels can help to treat different cancers like CLL [2, 3]. MicroRNAs (miRNAs), a novel class of endogenous small noncoding RNAs with 18-22 nucleotides, have been associated with diverse types of malignancies [4, 5]. By binding to the 3′-untranslated region (UTR) or 5´UTR of their mRNA targets, miRNAs control the gene expression at the post-transcriptional level, and lead to inhibition or degradation of mRNA. miRNAs play a major role in malignancies progression as oncogenes or tumor suppressors [6]. miRNAs have essential roles in cellular and molecular processes associated with CLL. Although miR-15a and miR-16-1 were discovered within the 13q14.3 deleted regions in CLL, the prognostic value of miRNAs still remains unknown [7]. Previous investigations has reported deregulated expression of miR-485-3p in several cancers [8-10]. However, one investigation by Di Lisio et al have reported involvement of miR-485-3p in CLL by assessing differentially expressed miRNAs in each sort of lymphoma [11]. In this study, the expression level of miR-485-3p was examined in peripheral blood mononuclear cells (PBMCs) of CLL patients in comparison with controls in order to determine whether deregulated expression of miRNA can be used as a diagnostic biomarker. Additionally, in silico analyses on miR-485-3p targetome indicated FOXD3 an involvement of target in both miR-485-3p and CLL disease. Thus, we also investigated FOXD3 expression by quantitative real-time polymerase chain reaction (PCR) in PBMCs of 35 CLL patients and the same number of controls.
Materials and Methods
In this case-control investigation, blood samples were collected from CLL patients and random samples from healthy men and women. Seventy samples, including 35 CLL patients [diagnosed in the Omid Hospital (Isfahan, Iran)] and 35 controls, were chosen for the current investigation. CLL patients were diagnosed by an expert on the basis of blood cell count and cell morphology. The Ethics Committee of Omid Hospital (Isfahan, Iran) approved the protocol of this study. Before taking a sample of individuals, a written informed consent was taken from each person. Four ml of peripheral blood was collected into ethylenediaminetetraacetate-containing tubes and was transferred to the laboratory on ice.
Complete blood count
White blood cell (WBC) counts, monocyte, and lymphocyte counts were assessed using CA&XN-Series TM Automated Hematology Analysis (Kobe, Japan).
Peripheral blood mononuclear cell extraction
The density gradient lymphoprep (Bio Sera, Kansas City, USA) was used to extract PBMCs from blood samples based on the manufacturer’s protocol. PBMCs were isolated from the middle phase into RNAase-free micro tubes and made frozen until RNA extraction step.
Total RNA isolation and complementary DNA synthesis
miRNA Hybrid-R (Geneall, Seoul, Korea) was applied to extract total RNA from PBMCs under RNase-free condition according to the manufacturer’s instruction. The quality of total RNAs was examined at a 260/280 nm wavelength ratio using NanoDrop spectrometer (Thermo Scientific, Waltham, MA, USA). cDNA synthesis for miR-485-3p and FOXD3 was fulfilled on a total RNA using a cDNA Synthesis Kit (Pars Genome, Tehran, Iran) according to the kit protocol. Then, the cDNA synthesized was stored at -20oC until the next step.
Real-time PCR
Relative expression of miR-485-3p and FOXD3 was investigated using real-time PCR analysis on the base of SYBR Green (TaKaRa, Kusatsu, Japan) exploration with Rotor-Gene 6000 system (Corbett Life Science, Australia). Glyceraldehyde-3-phosphate dehydrogenase (GAPDH) was selected as internal control, in order to normalize FOXD3 expression levels. Also, the final data for miR-485-3p were normalized using small nuclear RNA, U6, expression level as an endogenous control. The primers were procured from Bon Yakhteh (Tehran, Iran). 1 μl product of cDNA mixed to a master mix including 0.5 µl separate from each primer (forward and reverse), 3 µl of Diethylpyrocarbonate (DEPC)-treated water, and 5 μl of SYBR premix ExTaq II (TaKaRa, Kusatsu, Japan) in a total volume of 10 μl. The protocol was as follows: 95°C for 10 min, followed by 40 cycles at 95°C for 10 s, 60°C for 20 s and 72°C for 25 s. Triplicate experiments were applied to each sample.
Molecular enrichment analysis
In order to obtain suitable miRNA in CLL, vast search in the miRWalk 2.0 database was performed and miR-485-3p was selected. Subsequently, characterization of miR-485-3p targets was carried out by exploring at TargetScan v7 and some high-scoring miR-485-3p targets were selected. On the other hand, to find the associated gene with CLL disease, DisGeNET database was applied and genes associated with CLL were extracted from the database. Finally, a common gene target between miR-485-3p (TargetScan) and CLL (DisGeNET) was chosen. Furthermore, in silico molecular enrichment analysis was conducted on predicted and validated targets of miR-485-3p from the miRecords and miRTarBase databases, respectively [12-13].
Statistical analysis
Data obtained was performed by the 2-ΔΔCT statistical method.Data were analyzed using Graph Pad Prism statistical software, version 5.01 (Graph Pad, San Diego, CA, USA). Normality was evaluated by the Kolmogorov-Smirnov test. The independent samples t-test was applied to analyze the data between groups. In all tests, p≤0.05 was considered as the level of significance.
Results
Clinical features of study population
A study population including 35 CLL patients (average age: 62.05±1.85, range: 41-80 years, disease duration: 1.42±0.78 years) and 35 controls (average age: 47.05±1.85, range: 25-71 years) were enrolled. Control group had no history of the genetics of CLL and did not use any special medicine. The hematological parameters of CLL are shown in Table 1. It was observed that in CLL patients, the levels of WBCs, monocyte and lymphocyte counts, are higher than the normal values. p≤0.05 was considered significant. Data were expressed as mean±SD.
Up-regulation of miR-485-3p in CLL
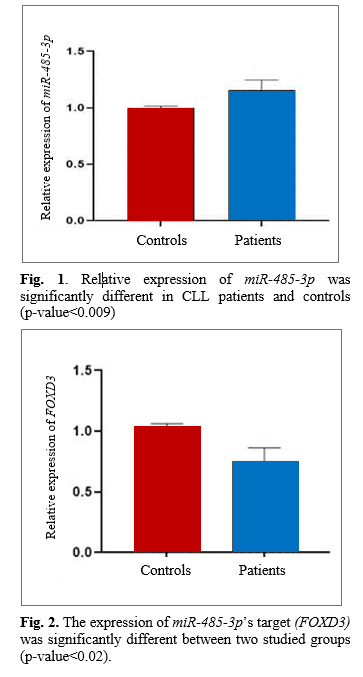
Data analysis indicated a significant increase in the expression of miR-485-3p in CLL patients compared with controls. The relative quantification (RQ) for the miR-485-3p was significantly different in both groups with a p <0.009 (Fig. 1). We observed a significant growth in the expression of miR-485-3p in CLL; therefore, it can be assumed that miRNA can be a potentially valuable biomarker for this disease.
Molecular enrichment analysis of miR-485-3p targetome
Using the miRecords database, 69 mRNAs were identified as predicted targets of miR-485-3p. All predicted targets were confirmed by at least five prediction databases. Similarly, using the miRTarBase database, 3 mRNAs were obtained as validated targets of miR-485-3p. The validated targets of miR-485-3p included NFYB, SLC40A1, and PBRM. Moreover, FOXD3 was selected as mentioned in the method section as miR-485-3p's target. Data analysis indicated a decreased expression of FOXD3 in CLL patients compared to the controls (p<0.02) (Fig. 2).
Table 1. Hematological parameters of chronic lymphocytic leukemia
| Characteristics |
Control |
Patient |
| Number of subjects |
35 |
35 |
| Mean number of white blood cells |
7.67 ± 0.39* |
65.23 ± 1.16 |
| Range (cells per mcL/103) |
4.5-9.9 |
6.03-451 |
| Mean number of monocytes |
0.453 ± 0.056 |
6.68 ± 2.01 |
| Range (cells per mcL/103) |
0.04-0.7 |
0.43-45.19 |
| Mean number of lymphocyte |
2.55 ± 0.177 |
44.93 ± 8.42 |
| Range (cells per mcL/103) |
1.2-3.7 |
2.38-408.03 |
* Data are presented as Mean±SD
Discussion
In the current study, we investigated the expression level of miR-485-3p by targeting FOXD3 using real-time PCR in CLL patients in an Iranian population. The results demonstrated that CLL patients have a significantly higher expression of miR-485-3p compared with the controls. In addition, we observed the expression level of miR-485-3p’s target (FOXD3) reducing significantly in CLL patients compared to the control. Previous investigations have indicated the prognostic prediction in CLL being based on V-gene mutations (CD38 and ZAP-70) and different sorts of chromosomal deletions, such as 17q13, 6q21, 11q23, and 13q14 in CLL [14]. CLL was almost the first human cancers found that was clearly associated influenced by changes in miRNA expression. Deletion of miR-15a and miR-16 in CLL was discovered to result in the up-regulation of BCL-2 gene [15]. Although the function of miR-485-3p in CLL is still unknown, in order to develop a miRNA-based biomarker for cancers or other diseases, one of the main prerequisites is the ability to quantify miRNAs from a diversity of samples with enough sensitivity. The relationship of miR-485-3p has been shown to CLL has only been identified in a previous microarray study. The expression of miR-485-3p to be higher in CLL patients than controls [11]. Therefore, in our study, miR-485-3p and its target (FOXD3) were selected from in silico databases as a miRNA which is involved in CLL. On the basis of the data analysis, overexpression of miR-485-3p may be of functional importance and be related to prognosis of CLL. It has been proposed that the combined inhibition of PI3K/Akt and PTEN activity may represent a novel concept for CLL therapy. On the other word, the PI3K/AKT pathway is the key signaling for various pathways involved in CLL pathogenesis and it is active in isolated CLL cells [16, 17]. On the other hand, it has been indicated that miR‑485 overexpression inhibits the activation of the AKT and ERK signalling pathways in glioblastoma cell lines. miR‑485 acts as a tumour suppressor in glioblastoma by targeting PAK4 and regulating the AKT and ERK signalling pathways [18]. Additionally, miR-485 suppresss the activity of PI3K/AKT/mTOR signaling in lung adenocarcinoma cells [19]. Smad4, Wnt1, and PI3K siRNAs in normal ESC culture conditions were transfected to determine the role of Smad, Wnt, and PI3K. Transfection of Smad4, Wnt1, and PI3K downregulated the expression of undifferentiation marker genes of ESCs, Oct4, Sox2, and FOXD3 [20]. A positive relationship between FoxD3 and AKT2 was discovered in HCC cells and tissues. Interestingly, Liu et al. reported that FOXD3 is down-regulated in hepatocellular carcinoma cells treated with AKT2 siRNA [21]. These findings suggest the relevance of miR-485-3p as an involved biomarker in CLL. It can be assumed that miR-485 may bear numerous target genes and the target genes may apply multiple purposes of miR-485-3p in biological processes. Overall, the findings of our study illustrats the communication of miR-485-3p and its target (FOXD3) to CLL.
Conclusion
The findings of this study were in accordance with what had previously been reported and therefore, it can be concluded that in our study population, too, miR-485-3p is upregulated and FOXD3 is downregulated in CLL patients. biomarker for the diagnosis of CLL.
Conflict of Interest
The authors declare no conflict of interest.
Acknowledgement
The authors would extend their deep gratitude to patients who participated in this investigation.
References
- Nateghi B, Behshood P, Fathullahzadeh S, Mardanshah O. Circulating miR-95 Is a potential biomarker of chronic lymphocytic leukemia. Res Mol Med. 2018; 26(1): 2017-2062.
- De Santis CE, Lin CC, Mariotto AB, Siegel RL, Stein KD, Kramer JL, et al. Cancer treatment and survivorship statistics. CA Cancer J Clin. 2014; 64(2): 252-71.
- Namazi F, Hadi N, Moghimi M, Eshaghiyan A, Nateghi B. Down-regulation of miR-193b-3p and miR-376a-3p in chronic lymphocytic leukemia. Res Mol Med. 2019; 19(64): 345-27.
- Fernando TR, Rodriguez Malave NI, Rao DS. MicroRNAs in B cell development and malignancy. J Hematol Oncol. 2012; 5(54): 7-23.
- Martini V, Frezzato F, Severin F, Raggi F, Trimarco V, Martinello L, et al. Abnormal regulation of BCR signalling by c‑Cbl in chronic lymphocytic leukaemia. Oncotarget. 2018; 9(32): 219-31.
- Reddy KB. MicroRNA (miRNA) in cancer. Cancer Cell Int. 2015; 15(2): 38-42.
- Nateghi B, Shams E, Behshood P, Fathullah-zadeh S, Salehi M. Expression profiles of miR-93 and miR-330 in iranian patients with chronic lymphocytic leukemia. Int J Med Lab. 2019; 6(2): 100-106
- Formosa A, Markert EK, Lena AM, Italiano D, Finazzi-Agro E, Levine AJ, et al. MicroRNAs, miR-154, miR-299-5p, miR-376a, miR-376c, miR-377, miR-381, miR-487b, miR-485-3p, miR-495 and miR-654-3p, mapped to the 14q32. 31 locus, regulate proliferation, apoptosis, migration and invasion in metastatic prostate cancer cells. Oncogene 2014; 33(44): 51-73.
- Lou C, Xiao M, Cheng S, Lu X, Jia S, Ren Y, et al. MiR-485-3p and miR-485-5p suppress breast cancer cell metastasis by inhibiting PGC-1α expression. Cell Death Dis. 2016; 7(3): 21-59.
- Chen Z, Li Q, Wang S, Zhang J. miR‑485‑5p inhibits bladder cancer metastasis by targeting HMGA2. Int J Mol Med. 2015; 36(4): 1136-142.
- Di Lisio L, Sánchez-Beato M, Gómez-López G, Rodríguez ME, Montes-Moreno S, Mollejo M, et al. MicroRNA signatures in B-cell lymphomas. Blood Cancer J. 2012; 57(2): 46-73.
- Xiao F, Zuo Z, Cai G, Kang S, Gao X, Li T. miRecords: an integrated resource for microRNA–target interactions. Nucle Acid Res. 2008; 37(1): 105-10.
- Hsu SD, Lin FM, Wu WY, Liang C, Huang WC, Chan WL, et al. miRTarBase: a database curates experimentally validated microRNA-target interactions. Nucleic Acids Res. 2010; 39(1): 163-69.
- Fathullahzadeh S, Mirzaei H, Honardoost MA, Sahebkar A, Salehi M. Circulating microRNA-192 as a diagnostic biomarker in human chronic lymphocytic leukemia. Cancer Gene Ther. 2016; 23(10): 32-7.
- Filip AA, Grenda A, Popek S, Koczkodaj D, Michalak Wojnowska M, Budzyński M, et al. Expression of circulating miRNAs associated with lymphocyte differentiation and activation in CLL-another piece in the puzzle. Ann Hematol. 2017; 96(1): 33-50.
- Shehata M, Schnabl S, Demirtas D, Hilgarth M, Hubmann R, Ponath E, et al. Reconstitution of PTEN activity by CK2 inhibitors and interference with the PI3-K/Akt cascade counteract the antiapoptotic effect of human stromal cells in chronic lymphocytic leukemia. Blood 2010; 116(14): 2513-521.
- Chapman CM, Sun X, Roschewski M, Aue G, Farooqui M, Stennett L, et al. ON 01910. Na is selectively cytotoxic for chronic lymphocytic leukemia cells through a dual mechanism of action involving PI3K/AKT inhibition and induction of oxidative stress. Clinic Cancer Res. 2012; 18(7): 1979-991.
- Mao K, Lei D, Zhang H, You C. MicroRNA-485 inhibits malignant biological behaviour of glioblastoma cells by directly targeting PAK4. Int J Oncol. 2017; 51(5): 1521-532.
- Mou X, Liu S. MiR-485 inhibits metastasis and EMT of lung adenocarcinoma by targeting Flot2. Biochem Biophysic Res Communic. 2016; 477(4): 521-26.
- Lee MY, Lim HW, Lee SH, Han HJ. Smad, PI3K/Akt, and Wnt‐dependent signaling pathways are involved in BMP‐4‐induced ESC self‐renewal. Stem Cell 2009; 27(8): 1858-68.
- Liu LL, Lu SX, Li M, Li LZ, Fu J, Hu W, et al. FoxD3-regulated microRNA-137 suppresses tumour growth and metastasis in human hepatocellular carcinoma by targeting AKT2. Oncotarget 2014; 5(13): 51-13.









































