Globally, lung cancer continues to be the major cause of cancer-related deaths among women and men. In 2018, the estimated number of new lung cancer cases and death were 11.6% and 18.4%, respectively [1]. This cancer is the second leading cause of cancer death among Iranian men and the third leading cause of cancer death among Iranian women [2]. Substantially, lung cancer incidence is tightly affected by cigarette smoking patterns; nearly 85% of lung cancer cases are caused by carcinogens in tobacco smoke [3]. Molecular heterogeneity is responsible for the critical problem in diagnosing and treating patients with lung cancer. Based on decades of study and research, lung cancer can be considered a multi-stage event, and the accumulation of genetic and epigenetic modifications leads to DNA damage and the transformation of normal lung epithelial cells into cancerous cells [4-6]. Non-small cell lung carcinoma (NSCLC) and small cell lung carcinoma (SCLC) are two main types of lung cancer in histological and clinical features. NSCLC accounts for about 85% of all types of lung cancer and divides into the subtypes of adenocarcinoma, squamous cell carcinoma, and large cell carcinoma [7]. Various treatment strategies have been used to treat this cancer, which target various events involved in cancer, such as angiogenesis and apoptosis. [8, 9]. Therefore, using treatment agents that can induce apoptosis is a very good idea in cancer therapy. Treatments for lung cancer include surgery, chemotherapy, radiotherapy, targeted therapy, or combination therapies. On the other hand, it is necessary to discover new chemotherapy drugs due to drug resistance [10, 11].
Natural compounds derived from animal, plant, and microbial sources have become popular in various sciences and industries, especially in drug production [12-14]. Microorganisms are the source of many important drugs, such as antibiotics, antitumor compounds, antiviral compounds, and antiparasitic agents [15-18]. Actinomycete strains are valuable and produce many natural bioactive products with antitumor properties [19]. It should be noted that the use of secondary metabolites of actinomycetes in various fields of pharmaceutical science and drug discovery, including the production of antitumor compounds, is promising and has attracted the attention of researchers [9, 20, 21]. Various studies on actinomycete strains have been performed in Iran, some of which aimed to identify actinomycetes that produce biologically active compounds. In 2014, Sarrami et al. proved the cytotoxic effects of some soil actinomycete strains stored at the University of Tehran. Microorganisms Collection (UTMC) against human lung cancer A549 cells [22]. However, isolated secondary metabolites of actinomycetes have not been studied to treat lung cancer by introducing apoptosis. Therefore, this study intended to evaluate the ability of actinomycete strains to induce apoptosis in A549 cells by measuring the quantitative expression of genes involved in apoptosis induction.
Materials and Methods
Cell culture
The human lung carcinoma cell line, A549, used in this study was obtained from the Iranian National Center for Genetic and Biologic Resources. A549 cells were plated in Dulbecco's Modified Eagle's medium (DMEM) (Gibco, USA) containing antibiotic (mixed 100 U/mL penicillin and 100 μmol/L streptomycins) (Gibco, USA) and 10% fetal bovine serum (FBS) (Gibco, USA) and maintained in a humidified incubator at 37 °C.
Microbial extract preparation
The secondary metabolites of UTMC 638 and UTMC 877 were obtained from the UTMC. The concentration of stock solutions prepared from each secondary metabolite was 4000 g/mL dissolved in dimethyl sulfoxide (DMSO) solution (Sigma, USA) [22]. The stock solutions were diluted with a culture medium to prepare the desired concentrations.
3-[4,5-dimethylthiazol-2-yl]-2,5 diphenyl tetrazolium bromide (MTT) assay
After treatment with UTMC 638, UTMC 877, and doxorubicin (Sigma, USA), the viability of A549 cells was evaluated by MTT assay (Sigma, USA). Briefly, 104 cells/well were plated in 96-well plates and exposed to different concentrations of UTMC 638 (0.8, 1.6, 3.2, and 6.4 µg/ml), UTMC 877 (6, 12, 24, and 48 µg/ml), and doxorubicin (0.125, 0.25, 0.5, and 1 µM) for 48 hours. In this study, doxorubicin was applied as a positive control. After the exposure, the medium was discarded from each well and replaced with a new medium containing MTT solution (1 mg/ml). After exposure to MTT, cells were incubated for 3 hours at 37 ˚C until a purple color of formazan product was visible [23]. The resulting formazan product was dissolved with DMSO, and the absorbance was detected at 570 nm using a microplate reader (BioTek, H1M).
Apoptosis detection via flow cytometry
The apoptosis rate of A549 cells was evaluated by annexin V-fluorescein isothiocyanate (FITC) and propidium iodide (PI) staining according to the manufacturer protocol (BD Biosciences, Clontech, USA). Shortly after treatment with UTMC 638, UTMC 877, and doxorubicin for 48 hours, A549 cells were trypsinized, rinsed, and then suspended in the Annexin-V binding buffer. Next, Annexin V-FITC and PI were added and incubated for 15 min at room temperature in the dark condition [24]. Finally, the rate of apoptotic cells was assessed by a flow cytometer using FlowJo software version 10 (FlowJo LLC, Ashland, OR, USA).
Real-Time polymerase chain reaction (PCR) Analysis
A real-time PCR technique was executed to determine the mRNA levels of the p53, Bax, p21, retinoblastoma, and caspase 7 genes. A549 cells were seeded in 6-well plates and exposed to UTMC 638, UTMC 877, and doxorubicin for 48 hours, and then total RNA isolation was performed using a total RNA isolation kit (CinnaGen, Iran) according to the manufacturer's instructions. One µg of total RNA was converted into the first-strand cDNA by First Strand cDNA Synthesis Kit (TAKARA, Japan).
Table 1. The specific sequences of selected primers used in this study
| Gene product |
Primer sequences |
Product size(bp) |
References |
| GAPDH |
Sense 5′- CCTCAAGATCATCAGCAATG-3′
Antisense 5′- CATCACGCCACAGTTTCC-3′ |
90 |
[26] |
| Bax |
Sense 5′-CAAACTGGTGCTCAAGGC-3′
Antisense 5′-CACAAAGATGGTCACGGTC-3′ |
178 |
[27] |
| Caspase-7 |
Sense 5′-CACGGTTCCAGGCTATTAC-3′
Antisense 5′-GGCAACTCTGTCATTCACC-3′ |
139 |
[27] |
| p21 |
Sense 5′-CCAGCATGACAGATTTCTACC -3′
Antisense 5′-AGACACACAAACTGAGACTAAGG-3′ |
150 |
[28] |
| P53 |
Sense 5′-GGAGTATTTGGATGACAGAAAC-3′
Antisense 5′-GATTACCACTGGAGTCTTC-3′ |
181 |
[29] |
| Retinoblastoma |
Sense 5′-AATCATTCGGGACTTCTG-3′
Antisense 5′-ACTTCCATCTGCTTCATC-3′ |
154 |
[29] |
Q-RT-PCR was conducted by RotorGene 6000 Q real-time analyzer (Corbett, Qiagen), and results were displayed by RotorGene 6000 Q real-time analyzer (Corbett, Qiagen). The sequences of the specific primers for p53, Bax, retinoblastoma, p21, Caspase7, and Glyceraldehyde-3-Phosphate Dehydrogenase (GAPDH), were given in Table 1. GAPDH was applied as an internal housekeeping control, and the selected mRNA expression was normalized to the GAPDH gene [25]. The present investigation results from a research project of the Department of Microbial Biotechnology University of Tehran through the following approved code: 1398/17.
Statistical analysis
All data were analyzed by GraphPad Prism software version 8 and presented as mean ± standard deviation (SD). Statistical differences were analyzed using one-way analysis of variance (ANOVA) followed by Tukey's multiple comparison test. P < 0.05 was considered a statistically significant difference.
Results
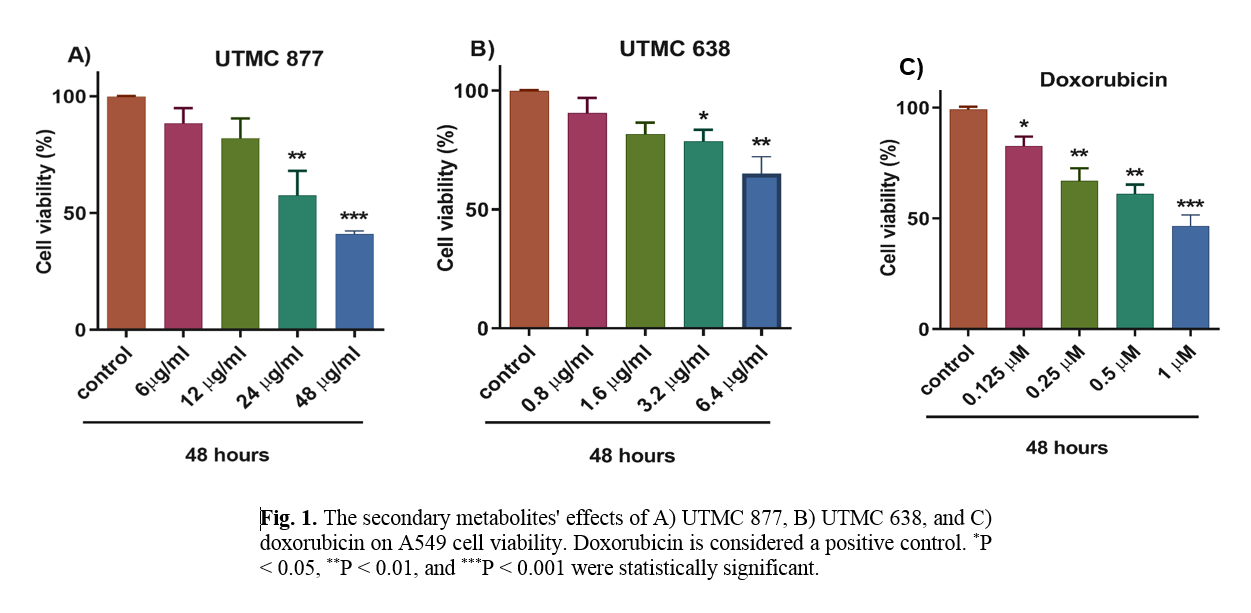
The effect of UTMC 638 and UTMC 877 secondary metabolites on A549 cell viability
As presented in Figure 1, data from the MTT assay showed that the crude extracts of UTMC 638, UTMC 877, and doxorubicin metabolites could reduce the viability of A549 cells in a dose-dependent manner. UTMC 877, UTMC 638, and doxorubicin at 24 µg/ml, 6.4 µg/ml, and 1 µM could decrease approximately 50% of A549 cell viability. These effective concentrations were used for subsequent analyses.
The effect of UTMC 638 and UTMC 877 secondary metabolites on morphological changes of A549 cells
As demonstrated in Figure 2, the lung cancer cells exposed to UTMC 638 and UTMC 877, as well as doxorubicin, morphologically exhibited cell death compared with the lung cancer cells exposed to DMSO (as a negative control).
The effect of UTMC 638 and UTMC 877 secondary metabolites on the apoptosis rate of A549 cells
Data from the flow cytometry indicated that UTMC 877, UTMC 638, and doxorubicin could promote apoptosis in the human lung cancer A549 cell line (Table 2, Figure 3). As depicted in Table 2, the necrotic and late apoptotic cell percentages in the A549 cells exposed to UTMC 877 were 47.3% and 17.7%, respectively, and in the cancer cells exposed to UTMC638 were 13.35% and 74.4%, respectively. However, after cell treatment with doxorubicin, the percentage of apoptotic and necrotic cells was about 38.35% and 37.45%, respectively. Furthermore, the late apoptotic rate in the A549 cells treated with UTMC 877 was notably lower than those exposed to doxorubicin or UTMC 638. However, the percentage of necrotic cells in UTMC 638-treated cells was lower than those treated with doxorubicin or UTMC 877 (Table 2).
The effect of UTMC 638 and UTMC 877 secondary metabolites on apoptosis-related gene expression
The quantitative analysis of retinoblastoma, Bax, p21, Caspase7 and p53 genes in cancer cells exposed to UTMC 877, UTMC 638, and doxorubicin was presented in Figure 4. The q-RT-PCR results showed that the expression of retinoblastoma, Bax, p21, Caspase7, and p53 genes were increased in the treated cells compared with control cells (p < 0.05, p < 0.01, p < 0.001).
Table 2. The impact of UTMC 877, UTMC 638, and doxorubicin on the apoptosis rate of A549 cells
| Group |
Flow cytometry (%) |
| Early apoptosis |
Late apoptosis |
Necrosis |
Alive |
| Control |
0.178 ± 0.007 |
2.905 ± 0.417 |
21.5 ± 3.111 |
75.4 ± 2.687 |
| UTMC 877 |
1.076 ± 0.585 |
17.70 ± 6.646 |
47.3 ± 3.67 |
34.25 ± 3.606 |
| UTMC 638 |
1 |
74.4 ± 8.83* |
13.35 ± 0.919 |
11.185 ± 6.809* |
| Doxorubicin |
1.755 ± 0.2* |
38.35 ± 8.697* |
37.45 ± 8.838 |
22.45 ± 0.07* |
All results are indicated at mean ± SD. *P < 0.05 was regarded as statistically significant.
.png)
.png)
Figure 4 shows that the mRNA expression of Bax, Casp7, and p21 genes was higher in the cells exposed to UTMC 638 than in the A549 cells treated with doxorubicin. The retinoblastoma gene expression in UTMC 638-treated cells was lower than in doxorubicin-treated cells (Figure 4). On the other hand, the mRNA expression of Casp7 and p21 genes was higher in the UTMC 877-treated cells than in doxorubicin-treated cells; however, the expression of the Bax and retinoblastoma genes in UTMC 877-treated cells was lower than doxorubicin-treated cells.
Discussion
In the present study, an NSCLC cell line, A549, was chosen as a cell model to investigate cell death and apoptosis induction in lung cancer cells. In the present study, we, for the first time, provided evidence using crude extracts UTMC 877 and UTMC 638 that two secondary metabolites of soil actinomycetes induce apoptosis in A549 cells. Our data indicated a decrease in the necrosis rate and an increase in the cell apoptosis rate after A549 cell incubation with UTMC 638 but not UTMC 877 secondary metabolites. The morphological examination also aligned with the results obtained from apoptosis in A549 cells.
The results of the present study were in line with the findings of previous studies on actinomycetes. According to previous information, actinomycetes were regarded as the major producers of new natural secondary metabolites with great pharmaceutical potential against chemotherapeutic-resistant tumors. Actinomycetes are an important resource of new anticancer metabolites, including metabolites displaying the capability to promote apoptosis [30, 31]. Elmallah et al. highlighted that marine actinomycetes-derived secondary metabolites could induce the downregulation of survivin and XIAP, leading to sensitization of MDA-MB-231 and HCT116 cells to cell death [32]. In another study, Zhou et al. reported that a natural product of marine actinomycete, Ilamycin E, induces apoptosis in triple-negative breast cancer cell lines HCC1937 and MDA-MB-468 [33]. Farnaes et al. found that napyradiomycin, as a derivative isolated by a marine-derived Actinomycete, induces cytotoxicity and apoptosis in the colon adenocarcinoma cell line HCT-116 [34].
Balachandran et al. proved that soil-obtained filamentous bacterium Streptomyces sp triggers apoptosis in A549 cells through p53, cytochrome c release, and caspase-dependent pathway [35]. Jeong et al. illustrated that Streptomyces sp. SY-103 metabolites induce apoptosis in human leukemia cells through activation of caspase-3 and inactivation of Akt [36]. Lin et al. revealed that marine actinomycetes-derived Actinomycin V alleviated the growth of A549 cells by inducing cell cycle arrest and apoptosis via the upregulation of p53, p21, and Bax genes. Therefore, this metabolite suppresses the growth of A549 cells by inducing p53 expression and promoting p53-dependent cellular events, including cell cycle arrest and apoptosis [37, 38]. In the present work, we also assessed the effect of UTMC 877 and UTMC 638 secondary metabolites on the expression of apoptosis-related genes. The gene expression analysis showed that UTMC 638 and UTMC 877, in line with apoptosis induction, effectively increase the expression of genes associated with cell apoptosis.
P53 is one of the classic tumor suppressor genes, which is frequently dysregulated in a large type of cancer cells. In normal cells, p53 contributes to transcriptional activation of downstream targets, including p21 and the pro-apoptotic proteins of BAX, PUMA, and NOXA [39]. The p21 protein is considered a universal inhibitor of cell cycle progression and exerts its functions on the cell cycle by inhibiting cyclin/CDK complexes and inactivating pRB [40]. The retinoblastoma and p53 genes are classified as tumor suppressors, preventing tumor formation and progression. This tumor suppressor protein regulates the cell cycle and several other biological functions. Retinoblastoma forms a transcriptional repression complex with the E2F transcription factor and inhibits the progression from G1 to S phase of the cell cycle by regulating E2F target genes [41]. Our findings collectively emerged that both UTMC 638 and UTMC 877 induced Bax, p21, p53, retinoblastoma, and Caspase7 mRNA expression. Consequently, apoptosis is raised, resulting in growth inhibition in A549 cells.
This study includes some limitations. Due to financial issues, the expression of the studied genes was executed only at the mRNA level, and their protein expression was not examined. Also, studying other genes involved in the intrinsic and extrinsic apoptosis pathways was impossible.
Conclusion
Our findings potentiate the idea that soil-derived secondary metabolites are probably introduced as novel pharmacological substances whose development may result in therapeutic pattern changes in oncology. The finding of current research highlighted that these metabolites could induce apoptosis in the human lung cancer cell line. However, these results may begin more extensive in vitro and in vivo studies.
Conflict of Interests
The authors declare that there is no conflict of interest associated with this work.
Acknowledgments
We would like to greatly acknowledge the Department of Microbial Biotechnology of the University of Tehran for its support. The present research was not supported by any funding organization of the commercial, nonprofit or public sectors.
References
- Sung H, Ferlay J, Siegel RL. Global cancer statistics 2020: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA: A Cancer Journal for Clinicians 2021; 71(3): 209-49.
- Khazaei S, Mansori K, Soheylizad M, Gholamaliee B, Khosravi Shadmani F, Khazaei Z, et al. Epidemiology of lung cancer in Iran: sex difference and geographical distribution. Middle East Journal of Cancer 2017; 8(4): 223-28.
- Shin DW, Kim YI, Kim SJ, Kim JS, Chong S, Park YS, et al. Lung cancer specialist physicians’ attitudes towards e-cigarettes: A nationwide survey. PloS one 2017; 12(2): 172568
- Testa U, Castelli G, Pelosi E. Lung cancers: molecular characterization, clonal heterogeneity and evolution, and cancer stem cells. Cancers 2018; 10(8): 248.
- Abbasi M, Mousavi MJ, Jamalzehi S, Alimohammadi R, Bezvan MH, Mohammadi H, et al. Strategies toward rheumatoid arthritis therapy; the old and the new. Journal of Cellular Physiology 2019; 234(7): 10018-10031.
- Pourgholi M, Abazari O, Pourgholi L, Ghasemi-Kasman M, Boroumand M. Association between rs3088440 (G> A) polymorphism at 9p21. 3 locus with the occurrence and severity of coronary artery disease in an Iranian population. Molecular Biology Reports 2021; 1(1): 1-8
- Zappa C, Mousa SA. Non-small cell lung cancer: current treatment and future advances. Translational Lung Cancer Research 2016; 5(3): 288
- Leprieur EG, Dumenil C, Julie C. Immuno-therapy revolutionises non-small-cell lung cancer therapy: results, perspectives and new challenges. European Journal of Cancer 2017; 78(1): 16-23.
- Panji M, Behmard V, Zare Z. Synergistic effects of green tea extract and paclitaxel in the induction of mitochondrial apoptosis in ovarian cancer cell lines. Gene 2021; 787(4): 145638.
- Johnson DH, Schiller JH, Bunn JPA. Recent clinical advances in lung cancer management. Journal of clinical oncology: official journal of the American Society of Clinical Oncology 2014; 32(10): 973-82.
- Abbasi M, Namjoo AR, Khamesipour F. Ethanol effects on histobiochemical parameters of suckling pups borned from alcoholic rat mothers. Comparative Clinical Pathology 2016; 25(4): 833-39.
- Ouyang L, Luo Y, Tian M. Plant natural products: from traditional compounds to new emerging drugs in cancer therapy. Cell proliferation 2014; 47(6): 506-15
- Huang CY, Ju DT, Chang CF, Reddy PM, Velmurugan BK. A review on the effects of current chemotherapy drugs and natural agents in treating non–small cell lung cancer. Biomedicine 2017; 7(4): 23.
- Mohamed R, Dayati P, Mehr RN. Transforming growth factor–β1 mediated CHST11 and CHSY1 mRNA expression is ROS dependent in vascular smooth muscle cells. Journal of cell Communi-cation and Signaling 2019; 13(2): 225-33.
- Łukasiewicz K, Fol M. Microorganisms in the treatment of cancer: advantages and limitations. Journal of Immunology Research 2018; 2397808.
- Singh M, Kumar A, Singh R, Pandey KD. Endophytic bacteria: a new source of bioactive compounds. Biotech. 2017; 7(5): 315.
- Asadi A, Nezhad DY, Javazm AR. In vitro effects of curcumin on transforming growth factor-β-mediated non-smad signaling pathway, oxidative stress, and pro‐inflammatory cytokines production with human vascular smooth muscle cells. Iranian Journal of Allergy, Asthma and Immunology 2020; 19(1): 84-93.
- Panji M, Behmard V, Zare Z, Malekpour M, Nejadbiglari H, Yavari S, et al. Suppressing effects of Green tea extract and Epigallocatechin-3-gallate (EGCG) on TGF-β-induced Epithelial-to-mesenchymal transition via ROS/Smad signaling in human cervical cancer cells. Gene 2021; 749: 145774.
- Woods GL, Elliott BA, Conville PS, Desmond EP, Hall GS, Lin G, et al. Susceptibility testing of mycobacteria, nocardiae, and other aerobic actinomycetes. 2nd ed. Wayne (PA): Clinical and Laboratory Standards Institute; 2011; 31(5): 24.
- Sharma M, Dangi P, Choudhary M. Actinomycetes: source, identification, and their applications. Int J Curr Microbiol App Sci. 2014; 3(2): 801-32.
- Dayati P, Rezaei HB, Sharifat N, Kamato D, Little PJ. G protein coupled receptors can transduce signals through carboxy terminal and linker region phosphorylation of Smad transcription factors. Life Sciences 2018; 199: 10-15.
- Sarrami S, Hamedi J, Mohammadipanah F, Rezayat Sorkhabadi SM. Study of cytotoxic effects of metabolites produced by actino-mycetes. Jundishapur Scientific Medical Journal 2014; 13(3): 347-55.
- Van Meerloo J, Kaspers GJ, Cloos J. Cell sensitivity assays: the MTT assay. Cancer cell culture: Springer, 2011; p. 237-45.
- Vermes I, Haanen C, Steffens-Nakken H, Reutellingsperger C. A novel assay for apoptosis flow cytometric detection of phosphatidylserine expression on early apoptotic cells using fluorescein labelled annexin V. Journal of Immunological Methods 1995; 184(1): 39-51.
- Schmittgen TD, Livak KJ. Analyzing real-time PCR data by the comparative CT method. Nature Protocols 2008; 3(6): 1101-108.
- Mondanizadeh M, Arefian E, Mosayebi G, Saidijam M, Khansarinejad B, Hashemi SM. MicroRNA‐124 regulates neuronal differ-entiation of mesenchymal stem cells by targeting Sp1 mRNA. Journal of Cellular Biochemistry 2015; 116(6): 943-53.
- Bohloli M, Atashi A, Soleimani M, Kaviani S, Anbarlou A. Investigating effects of acidic pH on proliferation, invasion and drug-induced apoptosis in lymphoblastic leukemia. Cancer Microenvironment 2016; 9(2-3): 119-26.
- Kazemi A, Sadri M, Houshmand M, Yazdi N, Zarif MN, Anjam-Najmedini A, et al. The anticancer effects of pharmacological inhibition of autophagy in acute erythroid leukemia cells. Anti-cancer Drugs 2018; 29(10): 944-55.
- Rezaie Z, Ardeshirylajimi A, Ashkezari MD. Improved anticancer properties of stem cells derived exosomes by prolonged release from PCL nanofibrous structure. Gene 2018; 665(1): 105-110.
- Guimarães TC, Gomes TS, Fernandes CD. Antitumor microbial products by actinomycetes isolated from different environments. Microbial Technology for Health and Environment 2020; 1(1): 113-60.
- Fattah A, Morovati A, Niknam Z, Mashouri L, Asadi A, Rizi ST, et al. The synergistic combination of cisplatin and piperine induces apoptosis in MCF-7 cell line. Iranian Journal of Public Health 2021; 50(5): 1037-47.
- Elmallah MI, Cogo S, Constantinescu AA, Elifio-Esposito S, Abdelfattah MS, Micheau O. Marine Actinomycetes-derived secondary metabolites overcome trail-resistance via the intrinsic pathway through downregulation of survivin and XIAP. Cells 2020; 9(8): 1760.
- Zhou W, Fang H, Wu Q, Wang X, Liu R, Li F, et al. Ilamycin E, a natural product of marine actinomycete, inhibits triple-negative breast cancer partially through ER stress-CHOP-Bcl-2. International Journal of Biological Sciences 2019; 15(8): 1723-732.
- Farnaes L, Coufal NG, Kauffman CA. Napyradiomycin derivatives, produced by a marine-derived actinomycete, illustrate cytotoxicity by induction of apoptosis. Journal of Natural Products 2014; 77(1): 15-21.
- Balachandran C, Sangeetha B, Duraipandiyan V, Raj MK, Ignacimuthu S, Al-Dhabi NA, et al. A flavonoid isolated from Streptomyces sp.(ERINLG-4) induces apoptosis in human lung cancer A549 cells through p53 and cytochrome c release caspase dependant pathway. Chemico-Biological Interactions 2014; 224(1): 24-35.
- Jeong SY, Han MH. Apoptosis induction of human leukemia cells by Streptomyces sp. SY-103 metabolites through activation of caspase-3 and inactivation of Akt. International Journal of Molecular Medicine 2010; 25(1): 31-40
- Lin SQ, Jia FJ, Zhang CY. Actinomycin V suppresses human non-small-cell lung carcinoma A549 cells by inducing G2/M phase arrest and apoptosis via the p53-dependent pathway. Marine Drugs 2019; 17(10): 572.
- Musavi H, Abazari O, Safaee MS. Mechanisms of COVID-19 entry into the cell: potential therapeutic approaches based on virus entry inhibition in COVID-19 Patients with underlying diseases. Iranian Journal of Allergy, Asthma, and Immunology 2021; 20(1): 11-23.
- Mirzayans R, Andrais B, Scott A, Murray D. New insights into p53 signaling and cancer cell response to DNA damage: implications for cancer therapy. Journal of Biomedicine and Biotechnology 2012; 2012: 170325.
- Xiao BD, Zhao YJ, Jia XY, Wu J, Wang YG, Huang F. Multifaceted p21 in carcinogenesis, stemness of tumor and tumor therapy. World Journal of Stem Cells 2020; 12(6): 481-87.
- Kitajima S, Li F, Takahashi C. Tumor milieu controlled by RB tumor suppressor. International Journal of Molecular Sciences 2020; 21(7): 2450.








































.png)
.png)
