Introduction
Urinary tract infection is one of the most common bacterial infections that occurs in all age groups, in which Escherichia coli (E. coli) is known as a significant etiologic factor in 50-80 percent of cases [1]. The severity of the urinary tract infections of this bacterium is due to the presence of a wide range of virulence factors. Presently, the general hypothesis is that pathogenic strains are derived from non-pathogenic strains due to horizontal gene transfer process. These strains contain various virulence factors (VFs) such as adhesions, toxins, siderophores and the like contributing. which contribute to the development of the infection procedure and each subsequently overcome the host immune system [2]. However, the various adhesion genes such as fimH, papC, iucC, sfa/foc are involved in virulence, colonization and invasion [3]. Isolates that harbor these genes can lead to the emergence of multi drug resistant (MDR) strains [4]. The global investigations indicate that the antibiotic resistance increases among urinary tract infections due to the variety of virulence factors, albeit usage of antimicrobial agents, microbial properties and therapeutic strategies are critical in this regard. The aim of this study was to recognize the frequency of various adhesion genes and also the antimicrobial susceptibility profile of collected E. coli strains from urinary tract samples.
Materials and Methods
Clinical samples and laboratory identification
In this cross-sectional study a total of 90 non-duplicative clinical isolates of Uropathogenic E.coli were collected over nine months from urinary tract infection (UTI) samples (defined as the presence of a positive urine culture ≥105 CFU/ml and pyuria) of patients admitted to Ayatollah Rouhani Hospital in Babol, north of Iran. The study was approved by the Ethics Committee of Babol University of Medical Sciences, Babol, Iran (No: MIBABOL.REC.1395.246). The isolates were confirmed by conventional biochemical, microbiological and API 20E test system (BioMerieux Inc, France). All strains were stored in liquid broth (Merck, Germany) containing 20% glycerol at -20°C for molecular usage.
Antibiotic susceptibility test
In accordance with the Clinical and Laboratory Standards Institute (CLSI document M100-S16) guidelines, the antimicrobial susceptibility profile was conducted on the Mueller-Hinton agar plates (Merck, Germany) using the standard disk diffusion method for following antimicrobial agents: Penicillin (10 μg), Nalidixic acid (30 μg), Cefixime (5 μg), Gentamicin (10 μg), Nitrofurantoin (300 μg), Amikacin (30 μg), Ciprofloxacin (5 μg), Imipenem (10 μg), Co-trimoxazole (25 μg), Cefotaxime (30 μg), Ceftazidime (30 μg) (Rosco, Denmark).
Polymerase chain reaction (PCR) method
DNA genomic was extracted by genomic DNA purification kit (Yekta-Tajhiz, Iran) as a recommendation protocol. Specific primer sequences used in this study are listed as below: sfa/foc (F:GGAGGAGTAATTACAAACCTGGCA, R: GAGAACTGCCCGGGTGCATACTCT) (410 bp), PapC (F: GACGGCACTGCTGCAGGGTGTGGCG, R: ATATCCTTTCTGCAGGGATGCAATA) (328 bp), fimH (F: TGCAGAACGGATCCGTGG, R: GCAGTCACCTGCCCTCCGGTA) (508 bp), and iucC (F: AAACCTGGCTTACGCAACTGT, R: ACCCGTCTGCAAATCATGGAT) (269 bp) (Bioneer, Korea) [3]. The total volume of the PCR reaction mixture was 25 μl, containing 1 μl of extracted template DNA, 2 μl of 10× PCR buffer, 0.6 μl MgCl2 (50 mM), 0.6 μl dNTPs (10 mM), 0.5 μl of each primers, 0.5 μl of Taq DNA polymerase (5 U/μl) (Amplicon Co., Denmark) and 19.3 μl double distilled water. Amplification was carried out in a Techne TC-512 thermocycler (Eppendorf, Hamburg, Germany) as follows: initial denaturation at 95°C for 5 min, 30 cycles of denaturation for 30s at 94°C, annealing for 30s at 57°C, extension for 60s at 72°C, and a final extension for 5 min. at 72°C. Finally 5 μl of each PCR product was electrophoresed at 80V for 1 hour in a 1% agarose gel, stained and visualized with ultraviolet illumination system (Bio-Rad, Hercules, USA).
Results
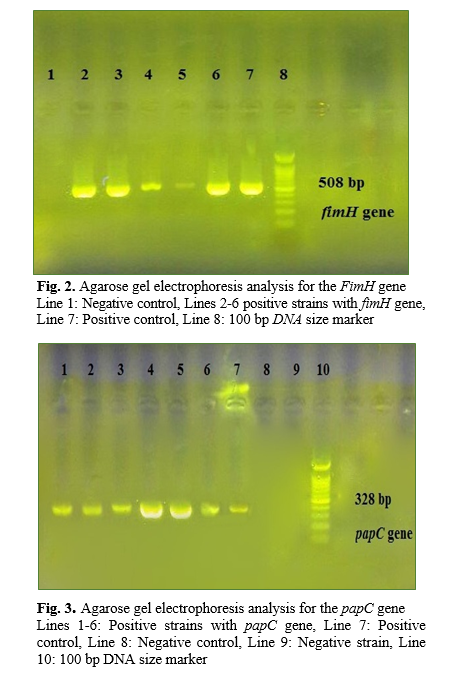
During the period of study, 90 UTI E. coli strains were obtained from different parts of Ayatollah Rouhani Hospital, (Babol, Iran) out of which 62.6% were above 50 years old with an average age of 55. Also, the patients’ gender comprised of 40% male and 60% female. The most percentage of the samples was isolated from infection disease (33.3%) and hematology-oncology wards (19%). The antimicrobial resistance pattern illustrated that penicillin with (100%) and imipenem by (0%) had the highest and lowest ratio. All examined antimicrobial agents are demonstrated in Fig 1. On the other hand, 43.3% of isolates were distinguished as MDR strains. By PCR molecular test the fimH with 66% had the highest rate among other genes while, the other genes were followed as: papC (46.7%), iucC (44.12%) and sfa/foc (39.4%). The PCR amplification of fimH, papC, iucC, and sfa/foc genes is presented in Figs 2-5.
According to some studies, the age of the patients may be an important risk factor in UTI, although in our results, the patients with more than 50 years old were sensitive to Uropathogenic E.coli infections [5, 6]. The antimicrobial susceptibility results were indicated that among tested antimicrobial agents, the imipenem (0%), amikacin (4.4%) and nitrofurantoin (15.5%) are the most effective antibiotics against Uropathogenic E.coli strains in the present study. Additionally, 43.3% of isolates were recognized as MDR strains. The prevalence of fimH gene (66%) may prove the mentioned issue in the present study for the reason that it can contain the attachment of E.coli to mucosal surfaces and initiate the UTI infection. Also, the similar studies have reported similar results around the high frequency of fimH gene in E.coli strains [7-9]. Moreover, the papC gene with 46.7%, which is responsible for the bacterial fimbriae function, has a significant role in attachment to eukaryotic cells [10]. Moreover, the iucC gene, which is responsible for the synthesis of the hydroxamate siderophore, and the other adhesion factor, sfa/foc gene, were determined 44.12% and 39.4% respectively. In López-Banda et al. study the prevalence of fimH and papC genes were 86.1% and 62% respectively [11]. However, in Tiba et al.'s study, the percentage of adhesion factors in patients with cystitis was reported as follows: fimH (97.5%), papC (32.7%), and iucC (25.9%) [12]. In the same study in Romania, the prevalence of fimH, sfa/foc and papC were 86%, 23% and 36%, respectively [13]. Chakraborty and colleagues (2015) demonstrated that the percentage of fimH and papC genes in 100 studied isolates amounted to 76% and 44%, respectively [14]. One of the differences between reported results may be related to the frequency of virulence factors of various strains and distribution of uropathogenic E.coli strains in various regions [15-18]. Moreover, in a current study by statistical analysis, there was a relationship between resistance to ciprofloxacin, gentamicin with papC, co-trimoxazole and fimH, iucC positive strains and resistance to cefixime (p-value <0.05). Furthermore, 30% of all strains harbored two genes, 20% of them harbored three genes and 11.1% had 4 positive genes simultaneously.
Conclusion
Uropathogenic E.coli is one of the main causes of UTI in different patients, hence the evaluation of the bacterial adhesion factors and the therapeutic approaches are the main aspects in UTI treatment. Due to an increase in the resistance genes in different conducted studies the dissemination of MDR strains is not unexpected.
Conflict of Interest
None to declare.
Acknowledgement
This study was funded by the research committee of Babol University of Medical Sciences, grant No.9543618 (4013).
References
[1]. Tarchouna M, Ferjani A, Ben-Selma W, virulence genes in Escherichia coli isolated from patients with urinary tract infection. Int J Infect Dis. 2013; 17(6): e450-3.
[2].
Dzidic S, Bedekovic V. Horizontal gene transfer-emerging multidrug resistance in hospital bacteria. Acta Pharmacol Sin. 2003; 24(6): 519-26.
[3]. Asadi S, Kargar M, Solhjoo K, Najafi A, Ghorbani-Dalini S. The association of virulence determinants of uropathogenic escherichia coli with antibiotic resistance. Jundishapur J Microbiol. 2014; 7(5): e9936.
[4]. Santo E, Salvador MM, Marin JM. Multidrug-resistant urinary tract isolates of Escherichia coli from Ribeirao Preto, Sao Paulo, Brazil. . 2007;(6): 575-8.
[5]. Chatterjee S, Todi S, Sahu S. Epidemiology of severe sepsis in India. Crit Care. 2009;.
[6]. - onset bacteremia due to extended-spectrum beta-lactamase- factors and prognosis. Clin Infect Dis. 2010; 50(1): 40-8.
[7]. Cooke NM, Smith SG, Kelleher M, Rogers TR. between community and nosocomial Escherichia coli bloodstream isolates. J Clin Microbiol. 2010; 48(4): 1099-104.
[8]. Mora A, López C, Dabhi G, Blanco M, Blanco JE, Alonso MP, et al. Extraintestinal pathogenic Escherichia coli O1:K1:H7/NM from human and avian origin: Detection of clonal groups host distribution. BMC Microbiol. 2009; 9(1): 132.
[9]. Ananias M, Yano T. Serogroups and virulence genotypes of Escherichia coli isolated from patients with sepsis. Braz J Med Biol Res. 2008; 41(10): 877-83.
[10]. Maynard C, Bekal S, Sanschagrin F, Levesque RC, Brousseau R, Masson L, et al. Heterogeneity among virulence and antimicrobial resistance gene profiles of extra intestinal Escherichia coli isolates of animal and human origin. J Clin Microbiol. 2004; 42(12): 5444-452.
[11]. López-Banda DA, Carrillo-Casas EM, Leyva-Leyva M, Orozco-Hoyuela G, Manjarrez-Hernández ÁH, Arroyo-Escalante S, et al. Biomed Res Int. 2014; 2014: 959206.
[12]. Tiba MR, Yano T, LeiteDda S. Genotypic characterization of virulence factors in Escherichia coli strains from patients with cystitis. Rev Inst Med Trop Sao Paulo. 2008; 50(2): 55-60.
[13]. Usein CR, Damian M, Tatu-Chitoiu D, Capusa C, Fagaras R, Tudorache D, et al. Prevalence of virulence genes in Escherichia coli strains isolated from Romanian adult urinary tract infection cases. J Cell Mol Med. 2001; 5(3): 303-10.
[14]. Genomic analysis and clinical importance of Escherichia coli isolate from patients with sepsis. Indian J Pathol Microbiol. 2015; 58(1): 22-6.
[15]. Santo E, Macedo C, Marin JM. Virulence factors of Uropathogenic Escherichia coli from a university hospital in Ribeirao Preto, Sao Paulo, Brazil. Rev Inst Med Trop Sao Paulo. 2006; 48(4): 185-88.
[16]. Blanco M, Blanco JE, Alonso MP, Mora A, Balsalobre C, Munoa F, et al. Detection of pap, sfa and afa adhesin-encoding operons
in Uropathogenic Escherichia coli strains: relationship with expression of adhesins and production of toxins. Res Microbiol. 1997; 148(9): 745-55.
[17]. Farshad S, Emamghorashi F. The prevalence of virulence genes of E. coli strains isolated from children with urinary tract infection. Saudi J Kidney Dis Transpl. 2009; 20(4): 613-17.
[18]. Benton J, Chawla J, Parry S, Stickler D. Virulence factors in Escherichia coli from urinary tract infections in patients with spinal injuries. J Hosp Infect. 1992; 22(2): 117-27.








































.PNG)
.PNG)
.PNG)
