Mon, Feb 2, 2026
[Archive]
Volume 6, Issue 2 (May 2019)
IJML 2019, 6(2): 100-106 |
Back to browse issues page
Download citation:
BibTeX | RIS | EndNote | Medlars | ProCite | Reference Manager | RefWorks
Send citation to:



BibTeX | RIS | EndNote | Medlars | ProCite | Reference Manager | RefWorks
Send citation to:
Nateghi B, Shams E, Behshood P, Fathullahzadeh S, Salehi M. Expression Profiles of miR-93 and miR-330 in Iranian Patients with Chronic Lymphocytic Leukemia. IJML 2019; 6 (2) :100-106
URL: http://ijml.ssu.ac.ir/article-1-256-en.html
URL: http://ijml.ssu.ac.ir/article-1-256-en.html
Department of Genetics and Molecular Biology, Faculty of Medicine, Isfahan University of Medical Sciences, Isfahan, Iran
Full-Text [PDF 342 kb]
(824 Downloads)
| Abstract (HTML) (3213 Views)
WBC= White blood cell; PLT= Platelet.
p≤0.05 was considered significant. Data were expressed as mean±SD.
.JPG)
Discussion
References
Full-Text: (1117 Views)
Introduction
Chronic lymphocytic leukemia (CLL) is a monoclonal disorder characterized by a progressive accumulation of functionally incompetent lymphocytes [1]. CLL is the most frequent type of leukemia recognized in western countries, accounting for about one-third of all cases of adult leukemias [2]. The incidence of this type of leukemia is 30 percent among all blood cancers and is slightly more common in men than in women [3]. The epidemiological evidence demonstrated that 5-10 percent of CLL cases are associated with a family history [4]. Advancing the understanding of CLL molecular genetics may have therapeutic stratification based on molecular prognosticators [5]. MicroRNAs (miRNAs), a new class of endogenous small noncoding RNAs with 18-22 nucleotides, have been associated with several types of cancer [6, 7]. miRNAs mediate post-transcriptional gene regulation, leading to the degradation or translation repression of target messenger RNAs (mRNAs) by recognization of 3' untranslated region of mRNAs [8]. In cancers, miRNAs can function as regulatory molecules or as oncogenes by down-regulating tumor suppressors [9]. The miR-15a and miR-16-1 were discovered in 2002 within the 13q14.3 deleted region in CLL [10]. miRNAs have pivotal roles in cellular and molecular processes related to CLL. However, the prognostic value of miRNAs is still unknown [11]. Some studies have shown the change in expression of miR-93 and miR-330 in several cancers including lung, prostate, and colon cancer [12-17]. Moreover, a previous study has suggested miR-93 and miR-330 as miRNAs related to major molecular genetic alterations in acute myeloid leukemia [18]. However, the molecular mechanisms of miR-93 and miR-330 in CLL are still unknown. In the current study, the expression of miR-93 and miR-330 was investigated in peripheral blood mononuclear cells (PBMCs) of CLL patients and controls by real-time PCR method for the first time. The aim was to determine whether the deregulated expression of miR-93 and miR-330 can be used as a diagnostic biomarker in CLL patients.
Materials and Methods
Totally, 60 samples including 30 patients diagnosed with CLL in the Omid Hospital (Isfahan, Iran) and 30 controls (both male and female) were selected for the study. CLL was diagnosed based on blood cell count, cell morphology, and clinical symptoms. The exclusion criteria were as follows: (i) CLL diagnosis more than 12 months before registration; (ii) Clinical Binet stage B or stage C; (iii) need for therapy according to the National Cancer Institute (NCI) guidelines [19]. Four ml peripheral blood was collected in EDTA-containing tubes and transported on ice to the laboratory. The study was approved by the Ethics Committee of the Medical Genetics Research Center of Genome. Written informed consent was received from all participating subjects prior to sample collection.
Complete blood count
Complete blood count (CBC) test was assessed using CA&XN-Series TM Automated Hemato-logy Analyses (Kobe, Japan). The Sysmex XN series made use of fluorescence flow cytometry. Some variables associated with CLL were determined by this device.
Peripheral blood mononuclear cells isolation
PBMCs were isolated from blood samples by density gradient lymphoprep (Bio Sera, Kansas City, USA) based on the manufacturer’s protocol. Mononuclear cells, monocytes, and lymphocytes have a lower density in comparison with erythrocytes and leukocytes; therefore, after centrifugation, they remain in an intermediate phase. Briefly, 4 ml of blood was diluted at a ratio of 1:1 with physiological saline and gradually added to the 4 ml lymphoprep solution gradient in a Falcon tube. The tubes were centrifuged at 800 ×g for 30 min, and then PBMCs were transferred from the middle phase into 2-mL RNAase-free microtubes and were frozen at -70°C until the next step.
miRNA extraction
miRNA from PBMCs was extracted using miRNA Hybrid-R (Geneall, Seoul, Korea) based on the manufacturer's instructions. The quality of miRNA was determined at a 260/280 nm wavelength ratio measured by a NanoDrop spectrometer (Thermo Scientific, Waltham, MA, USA).
Complementary DNA synthesis and real-time PCR
Complementary DNA (cDNA) synthesis was made using a standard kit (Pars Genome, Tehran, Iran) according to the manufacturer's leaflet. The real-time quantitative PCR reactions were carried out using a Rotor-Gene 6000 system (Corbett Life Science, Mortlake, Australia) in a total volume of 10 μl. Briefly, 20 ng μl −1 of cDNA product was added to a master mix, including 10 pmol μl −1 of miR-93, miR-330 and U6 (as a housekeeping gene) primers (Pars Genome) and 5 ml of SYBR premix ExTaq II (TaKaRa, Kusatsu, Shiga Prefecture, Japan). The conditions of PCR were as follows: 95°C for 15 min, 40 cycles of 95°C for 15 s, 60°C for 30 s, and 72°C for 30 s.
Statistical analysis
Real-time PCR data analysis was performed using the ΔΔCT method, where CT is the cycle threshold [20]. Data were then analyzed using SPSS software version 20 (Chicago, IL, USA). For all the tests, p ≤ 0.05 was considered as the level of significance.
Results
Clinical and biological features of the participants
In this study, two groups were sufficiently matched in terms of age and sex (P=0.11, 0.99; respectively). The demographic status of the participants were as follows: 30 patients with CLL (mean age: 60.87±1.59 years; range: 45-81 years; 18 males and 12 females; mean of disease duration: 1±0.67 years) and 30 healthy individuals (mean age: 44.03±1.95 years, range: 25-68 years, 21 males and 9 females) (Table 1). Moreover, it was observed that, the levels of some variables including white blood cells, lymphocyte, and platelet counts were associated with CLL and were significantly higher in the cases than in the controls.
Materials and Methods
Totally, 60 samples including 30 patients diagnosed with CLL in the Omid Hospital (Isfahan, Iran) and 30 controls (both male and female) were selected for the study. CLL was diagnosed based on blood cell count, cell morphology, and clinical symptoms. The exclusion criteria were as follows: (i) CLL diagnosis more than 12 months before registration; (ii) Clinical Binet stage B or stage C; (iii) need for therapy according to the National Cancer Institute (NCI) guidelines [19]. Four ml peripheral blood was collected in EDTA-containing tubes and transported on ice to the laboratory. The study was approved by the Ethics Committee of the Medical Genetics Research Center of Genome. Written informed consent was received from all participating subjects prior to sample collection.
Complete blood count
Complete blood count (CBC) test was assessed using CA&XN-Series TM Automated Hemato-logy Analyses (Kobe, Japan). The Sysmex XN series made use of fluorescence flow cytometry. Some variables associated with CLL were determined by this device.
Peripheral blood mononuclear cells isolation
PBMCs were isolated from blood samples by density gradient lymphoprep (Bio Sera, Kansas City, USA) based on the manufacturer’s protocol. Mononuclear cells, monocytes, and lymphocytes have a lower density in comparison with erythrocytes and leukocytes; therefore, after centrifugation, they remain in an intermediate phase. Briefly, 4 ml of blood was diluted at a ratio of 1:1 with physiological saline and gradually added to the 4 ml lymphoprep solution gradient in a Falcon tube. The tubes were centrifuged at 800 ×g for 30 min, and then PBMCs were transferred from the middle phase into 2-mL RNAase-free microtubes and were frozen at -70°C until the next step.
miRNA extraction
miRNA from PBMCs was extracted using miRNA Hybrid-R (Geneall, Seoul, Korea) based on the manufacturer's instructions. The quality of miRNA was determined at a 260/280 nm wavelength ratio measured by a NanoDrop spectrometer (Thermo Scientific, Waltham, MA, USA).
Complementary DNA synthesis and real-time PCR
Complementary DNA (cDNA) synthesis was made using a standard kit (Pars Genome, Tehran, Iran) according to the manufacturer's leaflet. The real-time quantitative PCR reactions were carried out using a Rotor-Gene 6000 system (Corbett Life Science, Mortlake, Australia) in a total volume of 10 μl. Briefly, 20 ng μl −1 of cDNA product was added to a master mix, including 10 pmol μl −1 of miR-93, miR-330 and U6 (as a housekeeping gene) primers (Pars Genome) and 5 ml of SYBR premix ExTaq II (TaKaRa, Kusatsu, Shiga Prefecture, Japan). The conditions of PCR were as follows: 95°C for 15 min, 40 cycles of 95°C for 15 s, 60°C for 30 s, and 72°C for 30 s.
Statistical analysis
Real-time PCR data analysis was performed using the ΔΔCT method, where CT is the cycle threshold [20]. Data were then analyzed using SPSS software version 20 (Chicago, IL, USA). For all the tests, p ≤ 0.05 was considered as the level of significance.
Results
Clinical and biological features of the participants
In this study, two groups were sufficiently matched in terms of age and sex (P=0.11, 0.99; respectively). The demographic status of the participants were as follows: 30 patients with CLL (mean age: 60.87±1.59 years; range: 45-81 years; 18 males and 12 females; mean of disease duration: 1±0.67 years) and 30 healthy individuals (mean age: 44.03±1.95 years, range: 25-68 years, 21 males and 9 females) (Table 1). Moreover, it was observed that, the levels of some variables including white blood cells, lymphocyte, and platelet counts were associated with CLL and were significantly higher in the cases than in the controls.
Table 1. Clinical and demographic characteristics of participants
| Characteristics | Control | CLL | P-value |
| Number of subjects | 30 | 30 | - |
| Sex: Number of males Number of females |
21 9 |
18 12 |
0.99 |
| Mean age | 44.03±1.95 | 60.87±1.59 | 0.11 |
| Range (years) | 25-68 | 45-81 | |
| Mean of disease duration (years) | - | 1 ± 0.67 | - |
| Range (years) | - | 0.5-2.5 | |
| Mean number of WBC | 7.077 ± 0.3239 | 63.87 ± 16.94 | 0.001 |
| Range (cells per mcL/103) | 4.5-9.9 | 6.03-451 | |
| Mean number of lymphocytes | 2.530 ± 0.1377 | 43.67 ± 15.34 | 0.009 |
| Range (cells per mcL/103) | 1.2-3.7 | 2.38-408.03 | |
| Mean numbdr of PLT | 302.3 ± 17.85 | 135.9 ± 8.493 | < 0.0001 |
| Range (mcL/103) | 155-420 | 21-273 |
p≤0.05 was considered significant. Data were expressed as mean±SD.
Relationship between the expression of miR-93 and miR-330 and CLL
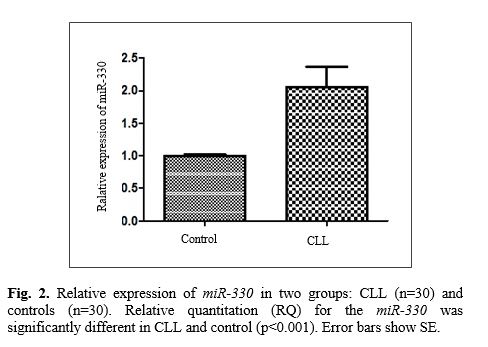
Data analysis indicated a significant increase in the expression of miR-93 in CLL patients compared with the controls. The relative quantitation (RQ) for the miR-93 was significantly different in CLL and controls with a p-value of <0.0001 (Fig. 1). Additionally, the results demonstrated significant growth in the expression of miR-330 in CLL patients compared with the controls (p<0.0015) (Fig. 2). We observed that the CLL patients were associated with higher levels of both miRNAs expression compared with the controls. Therefore, we hypothesized that this overexpression of miR-93 and miR-330 in CLL compared to the control could be studied as a potential therapeutic target to inhibit CLL progression in the future.
Data analysis indicated a significant increase in the expression of miR-93 in CLL patients compared with the controls. The relative quantitation (RQ) for the miR-93 was significantly different in CLL and controls with a p-value of <0.0001 (Fig. 1). Additionally, the results demonstrated significant growth in the expression of miR-330 in CLL patients compared with the controls (p<0.0015) (Fig. 2). We observed that the CLL patients were associated with higher levels of both miRNAs expression compared with the controls. Therefore, we hypothesized that this overexpression of miR-93 and miR-330 in CLL compared to the control could be studied as a potential therapeutic target to inhibit CLL progression in the future.
.JPG)

Discussion
In the present study, our results indicated that the patients had a significantly higher expression of miR-93 and miR-330 compared with controls. According to previous investiga-tions, prognostic prediction in CLL is based on V-gene mutations, CD38, ZAP-70, and deletion of the chromosome. The various types of chromosomal deletions, such as 17q13, 6q21, 11q23, 13q14 and 17p13, have been observed in CLL patients [21]. Appropriate non-aggressive biomarkers with high sensitivity and specificity are necessary for the diagnosis of the disease. miRNAs have emerged as biomarkers for the diagnosis and prognosis of different sorts of cancer. miRNAs have a deep impact on CLL development/progression [22-25]. Some studies have demonstrated the changes in expression of miR-93 in several cancers, including lung cancer, prostate cancer, and colon cancer [12-14]. Also, the biological role of miR‐330 has been reported in some cancers including lung cancer, prostate cancer, and colorectal cancer [15-17]. It has been suggested that miR-93 and miR-330 as a miRNAs are related to major molecular genetic alterations in acute myeloid leukemia [18]. However, as literature reveals, only one study has reported the involvement of miR-93 in CLL. Expression of miR-93 was significantly different in freshly isolated and cultured CLL cells carried out on a further group of patients using TaqMan real-time PCR [26]. In CLL, cancer cells originate from lymphoid cells, particularly B lymphocytes. These immature lymphocytes are observed in the blood. CLL is apperceived in lymph nodes, spleen, and other tissues [27]. The mechanisms for increased survival within tissues are not well understood but may provide novel targets for therapy [26]. Fabbri and Croce (2011) studied the role of microRNAs in lymphoid biology and disease. They suggested miR-93 (high expression) and miR-330 (high/low expression) as the most frequently de-regulated miRNAs in lymphoid malignancies [28]. In the current study, the association of miR-93 and miR-330 expression with CLL disease was analyzed by RT-qPCR. Our results revealed that CLL patients have a significantly higher expression of miR-93 and miR-330 compared with controls. The current investigation is the first study on the association of miR-93 and miR-330 expression with CLL carried out using RT-qPCR (as a gold standard method for gene expression analysis). The identification of genetic factors involving in CLL can be useful for better understanding of the prognosis and treatment of CLL. However, the use of miRNAs as a bio-marker still requires more investigatins.
Conclusion
According to the results of this study, miR-93 and miR-330 deregulation is probably associated with CLL and poor prognosis can have functional importance. Also, these miRNAs can be a potentially valuable marker for early diagnosis of CLL. The identification of the genetic factors involving in CLL can be useful for better understanding of the pathophysiology and prognosis of CLL.
Conflict of Interest
The authors declare no conflict of interest.
Acknowledgements
This study was supported by Isfahan University of Medical Sciences. We thank all patients who funded to this investigation.
Conclusion
According to the results of this study, miR-93 and miR-330 deregulation is probably associated with CLL and poor prognosis can have functional importance. Also, these miRNAs can be a potentially valuable marker for early diagnosis of CLL. The identification of the genetic factors involving in CLL can be useful for better understanding of the pathophysiology and prognosis of CLL.
Conflict of Interest
The authors declare no conflict of interest.
Acknowledgements
This study was supported by Isfahan University of Medical Sciences. We thank all patients who funded to this investigation.
References
- Hallek M. Chronic lymphocytic leukemia: 2013 update on diagnosis, risk stratification and treatment. Am J Hematol. 2013; 88(9): 803-16.
- Taghiloo S, Allahmoradi E, Tehrani M, Janbabaei G, Shekarriz R, Asgarian-Omran H. Blimp-1 expression as an exhaustion transcription factor in chronic lymphocytic leukemia. Res Mol Med. 2017; 15; 5(3): 5-10.
- Ahn IE, Underbayev C, Albitar A, Herman SE, Tian X, Maric I, et al. Clonal evolution leading to ibrutinib resistance in chronic lymphocytic leukemia. Blood 2017; 129(11): 1469-479.
- Ghia P, Ferreri AJ, Caligaris-Cappio F. Chronic lymphocytic leukemia. Critic Rev Oncol/ Hematol. 2007; 64(3): 234-46.
- Gaidano G, Foà R, Dalla-Favera R. Molecular pathogenesis of chronic lymphocytic leukemia. J Clinic Invest. 2012; 122(10): 3432-438.
- Bartel DP. MicroRNAs: genomics, biogenesis, mechanism, and function. Cell 2004; 116(2): 281-97.
- Ogata-Kawata H, Izumiya M, Kurioka D, Honma Y, Yamada Y, Furuta K, et al. Circulating exosomal microRNAs as biomarkers of colon cancer. PloS one 2014; 9(4): e92921.
- Mollainezhad H, Eskandari N, Pourazar A, Salehi M, Andalib A. Expression of microRNA-370 in human breast cancer compare with normal samples. Adv Biomed Res. 2016; 5(1): 129.
- Garzon R, Calin GA, Croce CM. MicroRNAs in Cancer. Annu Rev Med. 2009; 60(1): 167-79.
- Calin GA, Dumitru CD, Shimizu M, Bichi R, Zupo S, Noch E, et al. Frequent deletions and down-regulation of micro-RNA genes miR15 and miR16 at 13q14 in chronic lymphocytic leukemia. Proceed Nation Acad Sci. 2002; 99(24): 15524-5529.
- Mirzaei H, Fathullahzadeh S, Khanmohammadi R, Darijani M, Momeni F, Masoudifar A, et al. State of the art in microRNA as diagnostic and therapeutic biomarkers in chronic lymphocytic leukemia. J Cellul Physiol. 2018; 233(2): 888-900.
- Zhu W, He J, Chen D, Zhang B, Xu L, Ma H, et al. Expression of miR-29c, miR-93, and miR-429 as potential biomarkers for detection of early stage non-small lung cancer. PloS one 2014; 9(2): e87780.
- Choi N, Park J, Lee JS, Yoe J, Park GY, Kim E, et al. miR-93/miR-106b/miR-375-CIC-CRABP1: a novel regulatory axis in prostate cancer progression. Oncotarget. 2015; 15; 6(27): 23533.
- Yu XF, Zou J, Bao ZJ, Dong J. miR-93 suppresses proliferation and colony formation of human colon cancer stem cells. World J Gastroenterol. 2011; 17(42): 4711-717.
- Lee KH, Chen YL, Yeh SD, Hsiao M, Lin JT, Goan YG, et al. MicroRNA-330 acts as tumor suppressor and induces apoptosis of prostate cancer cells through E2F1-mediated suppression of Akt phosphorylation. Oncogene 2009; 28(38): 3360.
- Liu X, Shi H, Liu B, Li J, Liu Y, Yu B. miR-330-3p controls cell proliferation by targeting early growth response 2 in non-small-cell lung cancer. Acta Biochim Biophys Sin (Shanghai). 2015; 47(6): 431-40.
- Li Y, Zhu X, Xu W, Wang D, Yan J. miR-330 regulates the proliferation of colorectal cancer cells by targeting Cdc42. Biochem Biophys Res Communic. 2013; 431(3): 560-65.
- Babashah S, Sadeghizadeh M, Tavirani MR, Farivar S, Soleimani M. Aberrant microRNA expression and its implications in the pathogenesis of leukemias. Cellul Oncol. 2012; 35(5): 317-34.
- Negrini M, Cutrona G, Bassi C, Fabris S, Zagatti B, Colombo M, et al. microRNAome expression in chronic lymphocytic leukemia: comparison with normal B-cell subsets and correlations with prognostic and clinical parameters. Clin Cancer Res. 2014; 10(1): 723.
- Schmittgen TD, Livak KJ. Analyzing real-time PCR data by the comparative C T method. Nature Protocols 2008; 3(6): 1101.
- Fathullahzadeh S, Mirzaei H, Honardoost MA, Sahebkar A, Salehi M. Circulating microRNA-192 as a diagnostic biomarker in human chronic lymphocytic leukemia. Cancer Gene Therapy 2016; 23(10): 327.
- Mirzaei H, Gholamin S, Shahidsales S, Sahebkar A, Jaafari MR, Mirzaei HR, et al. MicroRNAs as potential diagnostic and prognostic biomarkers in melanoma. Euro J Cancer 2016; 53(1): 25-32.
- Salarinia R, Sahebkar A, Peyvandi M, Mirzaei HR, Jaafari MR, Riahi MM, et al. Epi-drugs and Epi-miRs: moving beyond current cancer therapies. Curr Cancer Drug Targets. 2016; 16(9): 773-788.
- Bartel DP. MicroRNAs: genomics, biogenesis, mechanism, and function. Cell 2004; 116(2): 281-97.
- Balatti V, Acunzo M, Pekarky Y, Croce CM. Novel mechanisms of regulation of miRNAs in CLL. Trends In Cancer 2016; 2(3): 134-43.
- Willimott S, Wagner SD. Stromal cells and CD40 ligand (CD154) alter the miRNome and induce miRNA clusters including, miR-125b/miR-99a/let-7c and miR-17-92 in chronic lymphocytic leukaemia. Leukemia 2012; 26(5): 1113.
- Li S, Moffett HF, Lu J, Werner L, Zhang H, Ritz J, et al. MicroRNA expression profiling identifies activated B cell status in chronic lymphocytic leukemia cells. PloS one 2011; 6(3): e16956.
- Fabbri M, Croce CM. Role of microRNAs in lymphoid biology and disease. Curr Opinion Hematol. 2011; 18(4): 266.
Type of Study: Research |
Subject:
Genetics/ Biotechnology
Received: 2018/09/1 | Accepted: 2019/05/11 | Published: 2019/05/31
Received: 2018/09/1 | Accepted: 2019/05/11 | Published: 2019/05/31
Send email to the article author
| Rights and permissions | |
 |
This work is licensed under a Creative Commons Attribution-NonCommercial 4.0 International License. |