Thu, Feb 19, 2026
[Archive]
Volume 7, Issue 2 (May 2020)
IJML 2020, 7(2): 110-120 |
Back to browse issues page
Download citation:
BibTeX | RIS | EndNote | Medlars | ProCite | Reference Manager | RefWorks
Send citation to:



BibTeX | RIS | EndNote | Medlars | ProCite | Reference Manager | RefWorks
Send citation to:
Jam Shahriari S, Alizadeh F, Khodavandi A. Interaction of Candida albicans with Fluconazole/ Clotrimazole: Effect on Hyphae Formation and Expression of Hyphal Wall Protein 1. IJML 2020; 7 (2) :110-120
URL: http://ijml.ssu.ac.ir/article-1-292-en.html
URL: http://ijml.ssu.ac.ir/article-1-292-en.html
Department of Microbiology, Yasooj Branch, Islamic Azad University, Yasooj, Iran.
Full-Text [PDF 505 kb]
(888 Downloads)
| Abstract (HTML) (1835 Views)
Introduction
Candida albicans (C. albicans) is a pleomorphic fungus that commonly exists as a normal resident microbiota of the human body. Nevertheless, this interaction between the host and microbe is not always benign, and as a pathogen, C. albicans is responsible for a wide range of infections from mucosal to invasive systemic candidiasis. However, pathogenesis of C. albicans is facilitated by environmental conditions [1-5]. Colonization of C. albicans occurs as a result of adherence to host epithelial cells [4, 6]. The adhesion of C. albicans to epithelial cells is a complex process and involve several different factors. Following adhesion of C. albicans to the cell surface, hyphae growing out of the yeast are thought to play a major role in the pathogenesis of the C. albicans and the infection progresses. There are a range of adhesins that bear the potential to interact with host epithelial cell receptors. One of the most studied adhesins to date is the hyphal wall protein 1 (HWP1). HWP1, which its N-terminal domain mimicing human epithelial cell transglutaminase substrates, is required for mucosal pathogenicity [2, 3, 7, 8]. HWP1 is strongly induced during hyphae growing out of the yeast. The transcriptional activators and repressors coordinately regulate HWP1 expression in C. albicans that control morphology [9, 10].
Much as considerable progress around antifungals has made adequate treatment of candidiasis, the available studies have demonstrated unacceptably high mortality associated with invasive systemic candidiasis [11-13]. For the treatment of candidiasis azole, antifungal agents are recommended. All of the azole antifungals inhibit the function of cytochrome P450 system to some degree of specificity [14]. The rapid emergence of azole drug-resistant of C. albicans has been reported [15, 16]. There are numerous evidences for effective combinations of antifungal agents [17, 18]. Mendling et al., revealed that combination of fluconazole (FLU) and clotrimazole (CLT) with the sequential dose of FLU is effective for the treatment of recurrent Candida vaginitis [19]. In the present study, we investigated the effect of combination of FLU/CLT on C. albicans hyphae formation. We have used gene expression profiling of hypha-specific gene (HWP1) to investigate the transcriptional responses of C. albicans hyphae exposed to combination of FLU/CLT at concentrations based on minimum inhibitory concentration (MIC).
Materials and Methods
C. albicans and chemicals
C. albicans ATCC 14053 reference strain was purchased from Iranian Research Organization for Science and Technology. Ten clinical isolates were obtained during recurrent vulvovaginal candidiasis infection (defined as 3 or more per year) in patients who had previously used FLU and CLT in the past 5 years. Frozen glycerol stock of the isolates were regularly revived on Sabouraud Dextrose Agar (SDA, Merck Research Laboratories, Darmstadt, Germany). Roswell Park Memorial Institute (RPMI)-1640 medium with L-glutamine without sodium bicarbonate (Sigma-Aldrich Co. St. Louis, MO, USA) was buffered with 0.165 M morpholine-propanesulfonic acid (MOPS, Sigma-Aldrich) to a pH of 7. Stock solutions of FLU (Sigma-Aldrich) and CLT (Sigma-Aldrich) were prepared in dimethyl sulfoxide (DMSO, Sigma-Aldrich) according to recommendations provided by the Clinical and Laboratory Standards Institute (CLSI) M27-A3 guidelines and stored at -20˚C until use. The FLU and CLT were mixed in 1:1 ratio [20, 21].
Interaction of fluconazole/clotrimazole
The interaction between FLU and CLT was determined on the basis of the fractional inhibitory concentration (FIC) index. The FIC index was defined as follows: [(MIC of drug A in combination/MIC of drug A alone)] + [(MIC of drug B in combination/MIC of drug B alone)]. The interaction was defined as synergistic if the FIC index was ≤0.5, partial synergy when the FIC> 0.5 but < 1.0, additive if the FIC = 1.0, indifferent when the FIC index > 1.0 but < 4.0, and antagonistic if the FIC ≥ 4.0 [22].
Time-killing test
In order to further confirm the effect of FLU and CLT alone and in combination, C. albicans ATCC 14053 and CI-5- isolate were used to determine the time-killing curves. Initial inoculum at a density of 1×106 cells/ml were treated with different concentrations (2×MIC and 1×MIC) of FLU and CLT alone and in combination. The number of viable cells was determined by colony counting at 0, 2, 4, 6, 8, 10, 12, 24, and 48 h after exposure to antifungal agents at 35˚C. The interaction between FLU and CLT tested by time-kill methods (at 24 to 48 h) were determined as follows: synergy, ≥2 log10 decrease in CFU/ml compared to the most active agent; additive effect <2 but >1 log10 decrease in CFU/ml compared to the most active agent; indifference, <2 but >1 log10 increase in CFU/ml compared to the least active agent; and antagonism, a ≥2 log10 increase in CFU/ml compare to the least active agent [23, 24].
Combination of fluconazole/clotrimazole on C. albicans hyphae
The hyphae formation assay was performed in U-bottomed 96-well polystyrene microtiter plate as described previously [25]. Briefly, 100 μl of the C. albicans ATCC 14053 cell suspension (1×106 cells/ml) was dispensed into the wells of microtiter plates. Different concentrations (2×MIC, 1×MIC, ½×MIC and ¼×MIC) of FLU and CLT alone and in combination were added (100 μl per well) to each well and incubated at 35◦C for 90 min without agitation. The microtiter plate was incubated at 35◦C for 16 h with gentle shaking. The metabolic activity of the C. albicans hyphae was determined quantitatively using colorimetric XTT [2,3-bis (2-methoxy-4-nitro-5 sulfophenyl)-5-[(phenylamino) carbonyl 2]-H-tetrazolium hydroxide] reduction and crystal violet assays.
XTT reduction assay of C. albicans hyphae
At the end of appropriate incubation, the supernatant was aspirated from the wells and washed 3-times with sterile phosphate-buffered saline. The C. albicans cell viability was determined using colorimetric XTT reduction assay [25]. Sterilized XTT/menadione (Sigma-Aldrich) solution transferred to each well containing prewashed hypha and incubated in the dark at 37◦C for 5 h. After the incubation, colorimetric change in the XTT reduction was measured in a microtiter microplate reader at 490 nm.
Crystal violet assay of C. albicans hyphae
The prewashed C. albicans hyphae was fixed with methanol and air dried. Then, C. albicans hyphae was stained with crystal violet solution (Sigma-Aldrich) for 20 min. Hundred μl of acetic acid 33% (Sigma-Aldrich) was distributed into the wells containing prewashed hyphae. The optical density was measured at 590 nm with Stat Fax 303 Reader (Awareness Technology) [25-27].
Scanning electron microscopy of C. albicans hyphae
Standard cell suspension of C. albicans was diluted (1×106 cells/ml) in RPMI-1640 medium supplemented with combination of FLU/CLT at MIC concentration on ThermanoxTM plastic coverslips (Nunc, Denmark) in 6-well cell culture plates (Nunc). The cell culture plates were incubated at 35◦C for 90 min (without agitation) and incubated again at 35◦ C for 16 h with gentle shaking. Thereafter, the prewashed hyphae was fixed in 2% (v/v) glutaraldehyde in phosphate-buffered saline (PH 7.2). Samples were washed with sodium cacodylate buffer and placed in 1% osmium tetroxide for 2 h at 4◦ C.
Samples were subsequently washed with sodium cacodylate buffer, dehydrated in a series of ascending ethanol solutions, put into critical point dryer and then stuck onto the stub. The specimens were coated with gold and viewed with a Philips XL30 (ESEM, UK) scanning electron microscope [25].
qRT-PCR analysis of C. albicans hypha-specific gene
C. albicans hyphae was performed in the presence of standard cell suspension of C. albicans and different concentrations (2×MIC and 1×MIC) of FLU and CLT alone and in combination in 6-well cell culture plates (Nunc).
Total RNA was extracted from C. albicans hyphae using RNeasy Mini Kit (Qiagen, Hilden, Germany) and 0.5 µg of total RNA was reverse transcribed with Moloney-Murine Leukemia Virus (MMLV) reverse transcriptase and random hexamers (Fermentas, USA). Resultant cDNAs were amplified by real time polymerase chain reaction (RT-PCR) using hypha specific and housekeeping internal control beta-actin gene primers (Table 1). Real time PCR reactions were run with ™SYBR Green qPCR Master Mix (Fermentas, USA) on Bio-Rad MiniOpticonTM system (USA). Relative expression was quantified by the Pfaffl method [23, 25].
The Research Ethics Committees of our institute (Ethical code 1213469) approved the study. The study protocol conformed to the ethical guidelines of the 2013 Declaration of Helsinki. Informed consent was obtained from patients.
Statistical analysis
Data were expressed as mean values of the biological replicates±standard deviations. One-way analysis of variance was applied to test the differences between treated and control groups. Tukey's HSD test was performed for a multiple comparison and statistical significance was tested at the p<0.05 levels. Statistical analyses were performed using the software SPSS 21.0 for windows (SPSS Inc. Chicago, IL, USA).
Results
Table 2 illustrates the susceptibility of clinical isolates of C. albicans and reference strains of C. albicans ATCC 14053 to FLU alone and in combination with CLT. The isolates were considered resistant at the MIC value ≥8.0 μg/ml for fluconazole, and ≥1.0 μg/ml for CLT as established by the CLSI M27-A3. As presented in table 2, all clinical isolates of C. albicans were resistant to FLU (MIC range 12.30-18.90 μg/ml), and CLT (MIC range 1.25-2.50 μg/ml).
The combined effects of the FLU and CLT against clinical isolates of C. albicans and reference strains of C. albicans ATCC 14053 are shown in table 3. A comparison between these values showed that the combination of FLU and CLT can be decreased from its MIC value of FLU and CLT by 6.79- to 61.50-fold and 1.04- to 6.25-fold, respectively. FLU showed synergistic (70%), partial synergistic (20%) and indifferent (10%) interaction with CLT against clinical isolates of C. albicans.The synergism of FLU/CLT against C. albicans ATCC 14053 and CI-5 isolate were confirmed by time-killing curves (Fig. 1). The combination of FLU/CLT at 2×MIC and 1×MIC concentrations against C. albicans ATCC 14053 revealed a 4.06, 4.10, 3.16 and 3.60 log10 CFU /ml decrease compared with FLU alone at 24 and 48 h, respectively. The results indicated that FLU and CLT action alone have little antifungal effect throughout 48 h in CI-5- isolate of C. albicans, but the combination yielded 2 × MIC and 1 × MIC a 6.03, 4.80, 7.02 and 5.95 log10 CFU/ml decrease compared with fluconazole alone at 24 and 48 h, respectively.
CI= Clinical isolates of C. albicans
CI= Clinical isolates of C. albicans
Table 4 exhibits the significant results to reduce hyphae after treatment with antifungal agents using XTT and CV assays for FLU and CLT alone and in combination in all concentrations based on MIC (p=0.001). XTT and CV assays for hyphae quantification also indicated C. albicans ATCC 14053 hyphae treated with combination of FLU and CLT being more significant than FLU and CLT alone (p=0.001).
The combination of FLU and CLT against C. albicans ATCC 14053 hyphae was visually verified by scanning electron microscopy (SEM) (Fig. 2). The growing hyphae was composed by yeast cells and hyphae form in untreated control group. The FLU and CLT combination-treated hyphae reduced in number and density of cells and also completely destroyed C. albicans hyphae.
In qRT-PCR, a variable Ct value of HWP1 gene in C. albicans ATCC 14053 hyphae treated with FLU and CLT alone and in combination revealed their gene expression levels being affected by the treatments (Fig. 3). The mean Ct values revealed that the combination of FLU/CLT induces a significant decrease in expression level of the HWP1 gene by 2.86- and 2.33-fold at 2× MIC and 1× MIC concentrations, respectively (p=0.002). Furthermore, the expression of HWP1 gene in C. albicans ATCC 14053 hyphae treated with FLU and CLT FLU and CLT alone showed a variable level of expression compared to the untreated control.
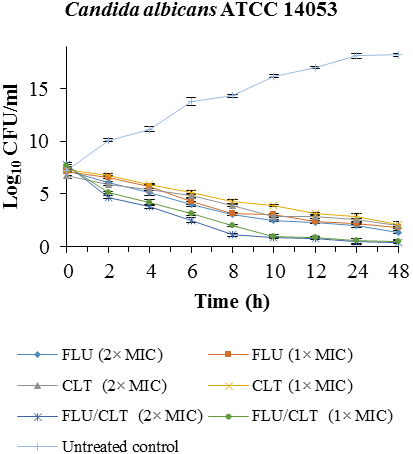

Fig. 1. Time-killing curves of FLU and CLT alone and in combination against C. albicans ATCC 14053 and CI-5 isolate at 0, 2, 4, 6, 8, 10, 12, 24, and 48 h. Data is represented as mean±SD.

Fig. 2. Scanning electron microscopy of C. albicans ATCC 14053 hyphae. (A) Untreated control, the morphology is composed of yeast cells and hyphae form. (B) Treated with combination of FLU and CLT at MIC concentration, complete destruction of hyphae form and reduced yeast cells were observed. Magnification 1250×.
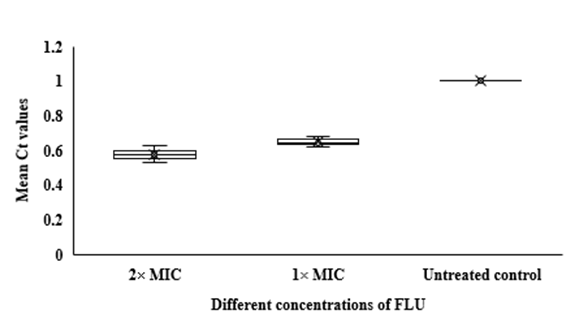

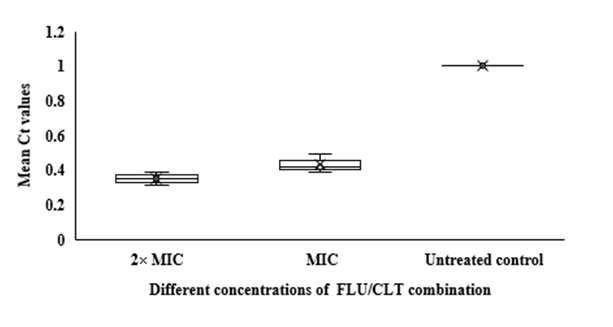
Fig 3. Whisker box plots of HWP1 gene expression at different concentrations of FLU and CLT alone and in combination based on MIC in C. albicans ATCC 14053 hyphae. The boxes represent the mean Ct values and the line inside the box represents median values. The bars across the boxes show locations of the minimum and maximum Ct values. Whiskers in the plot represent 95% confidence intervals. Data represent mean values±SD.
Discussion
Therapeutic strategies for fungal infections are limited by the small number of available antifungal drugs so that there is a need for efficient methods to identify novel drug targets and develop effective drug combinations [23, 30]. The switch between yeast and hyphae is a major role in the pathogenesis of C. albicans that could serve as target for treatment of infections. Several studies have proven useful in achieving that goal [23, 30-32]. In the current report, we demonstrate the combination of FLU/CLT on C. albicans and uncover their effective combinations on hyphae formation.
A synergistic effect was observed when FLU combined with CLT in clinical isolates proved resistant to FLU and CLT. FLU and CLT alone and in combination inhibited hyphal formation; they inhibited cell proliferation at higher concentrations. Combination of FLU/CLT is an extensively investigated drug combination that was shown to inhibit Candida cells [19, 33]. In an attempt to investigate the effectiveness combination of FLU/CLT on C. albicans hyphae, the growing hyphae was verified by XTT and CV assays and SEM. The combination of FLU and CLT destroyed hyphae and yeast form, suggesting their effective combinations on hyphae formation. We then analysed the expression level of hyphae specific gene in the C. albicans hyphae. Of three treatments, combination of FLU/CLT decreased and more variable expression of hyphae specific gene; in addition, FLU and CLT alone demonstrated a variable level of expression thus suggesting a change in the expression of HWP1 gene involved in hyphae formation and pathogenesis. We confirmed that hyphae is a target of the combination of FLU/CLT by showing inhibition of cell proliferation and hyphae formation and molecular analysis of hyphae specific gene in C. albicans.
Microbial analysis has already indicated that azole antifungal drugs are effective on C. albicans hyphae formation [23, 31, 34, 35]. Likewise, a role for HWP1 in C. albicans hyphal growth had been demonstrated by adhesins to host epithelial cell receptors [2, 3, 7, 8]. Our study provides further support for this interaction by linking pharmacological inhibition of hyphae formation. The inhibition of hyphae formation was accompanied by a reduction in hyphae specific gene expression. Khodavandi et al. exhibited that combination of FLU/ amphotericin B reduced yeast-hyphal transition and biomass and metabolic activity of the hypha in C. albicans [23]. Wakabayashi et al. demonstrated that triazole antifungal agents combined with triazoles lactoferrin-related compounds inhibit the growth of hyphae, an important form of pathogenesis in azole-resistant C. albicans strains [30]. The synergistic antifungal effect of licofelone in combination with fluconazole against C. albicans strongly reduced hyphal formation. The expression of RAS/cAMP/PKA pathway related genes (RAS1, CYR1, TPK2), and biofilm formation related genes including biofilm and cell wall regulator 1 (BCR1), HWP1, agglutinin-like sequence (ALS) 1 and ALS3 genes were down-regulated with the presence of licofelone combined with fluconazole [31]. FLU in combination with CLT revealed significant synergistic effects against C. tropicalis with FIC indices ranging from 0.011 to 0.43. In addition, the combination of FLU with CLT could trigger a down-regulation of C. tropicalis agglutinin-like sequence (ALS1 and ALS3), lipase (LIP1 and LIP4), and secreted aspartyl protease (SAP2 and SAP4) genes, respectively [36].
Conclusion
Our intention in assaying the effectiveness combination of FLU/CLT on C. albicans was to investigate the hyphae formation and transcriptional responses. Our results suggest that the HWP1 might constitute useful targets for the combination of FLU and CLT. This raises the prospect of identifying molecular mechanisms of combination of FLU/CLT as potential inhibitors of C. albicans virulence. However, this study demonstrated that the antifungal effect of FLU in combination with CLT could be critical in developing therapeutic strategies against C. albicans.
Conflict of interest
The authors declared no conflict of interest.
Acknowledgement
This research was supported by Islamic Azad University of Yasooj, Yasooj, Iran.
References
Full-Text: (1498 Views)
Introduction
Candida albicans (C. albicans) is a pleomorphic fungus that commonly exists as a normal resident microbiota of the human body. Nevertheless, this interaction between the host and microbe is not always benign, and as a pathogen, C. albicans is responsible for a wide range of infections from mucosal to invasive systemic candidiasis. However, pathogenesis of C. albicans is facilitated by environmental conditions [1-5]. Colonization of C. albicans occurs as a result of adherence to host epithelial cells [4, 6]. The adhesion of C. albicans to epithelial cells is a complex process and involve several different factors. Following adhesion of C. albicans to the cell surface, hyphae growing out of the yeast are thought to play a major role in the pathogenesis of the C. albicans and the infection progresses. There are a range of adhesins that bear the potential to interact with host epithelial cell receptors. One of the most studied adhesins to date is the hyphal wall protein 1 (HWP1). HWP1, which its N-terminal domain mimicing human epithelial cell transglutaminase substrates, is required for mucosal pathogenicity [2, 3, 7, 8]. HWP1 is strongly induced during hyphae growing out of the yeast. The transcriptional activators and repressors coordinately regulate HWP1 expression in C. albicans that control morphology [9, 10].
Much as considerable progress around antifungals has made adequate treatment of candidiasis, the available studies have demonstrated unacceptably high mortality associated with invasive systemic candidiasis [11-13]. For the treatment of candidiasis azole, antifungal agents are recommended. All of the azole antifungals inhibit the function of cytochrome P450 system to some degree of specificity [14]. The rapid emergence of azole drug-resistant of C. albicans has been reported [15, 16]. There are numerous evidences for effective combinations of antifungal agents [17, 18]. Mendling et al., revealed that combination of fluconazole (FLU) and clotrimazole (CLT) with the sequential dose of FLU is effective for the treatment of recurrent Candida vaginitis [19]. In the present study, we investigated the effect of combination of FLU/CLT on C. albicans hyphae formation. We have used gene expression profiling of hypha-specific gene (HWP1) to investigate the transcriptional responses of C. albicans hyphae exposed to combination of FLU/CLT at concentrations based on minimum inhibitory concentration (MIC).
Materials and Methods
C. albicans and chemicals
C. albicans ATCC 14053 reference strain was purchased from Iranian Research Organization for Science and Technology. Ten clinical isolates were obtained during recurrent vulvovaginal candidiasis infection (defined as 3 or more per year) in patients who had previously used FLU and CLT in the past 5 years. Frozen glycerol stock of the isolates were regularly revived on Sabouraud Dextrose Agar (SDA, Merck Research Laboratories, Darmstadt, Germany). Roswell Park Memorial Institute (RPMI)-1640 medium with L-glutamine without sodium bicarbonate (Sigma-Aldrich Co. St. Louis, MO, USA) was buffered with 0.165 M morpholine-propanesulfonic acid (MOPS, Sigma-Aldrich) to a pH of 7. Stock solutions of FLU (Sigma-Aldrich) and CLT (Sigma-Aldrich) were prepared in dimethyl sulfoxide (DMSO, Sigma-Aldrich) according to recommendations provided by the Clinical and Laboratory Standards Institute (CLSI) M27-A3 guidelines and stored at -20˚C until use. The FLU and CLT were mixed in 1:1 ratio [20, 21].
Susceptibility testing
Antifungal activity of FLU and CLT alone and in combination against planktonic cells of clinical isolates of C. albicans was tested by broth microdilution method using CLSI M27-A3 guidelines. Serially double-diluted concentrations of FLU range 0.0312–64 μg/ml and CLT range 0.0313-16 μg/ml alone and in combination were prepared in RPMI-1640 medium. One hundred μl of each dilution was dispensed into the well of a presterilized, U-bottomed 96-well polystyrene microtiter plate in the presence of 100 μl of the C. albicans planktonic cell suspensions of final density 5×102- 2.5×103 cells/ml. RPMI-1640 medium containing 5% DMSO was included in control wells. Thereafter, microtiter plates were kept at 4◦ C for 2 h and incubated at 35◦ C for 24 h. After incubation, growth of cells was measured by Stat Fax 303 Reader (Awareness Technology, Inc., USA) at a wavelength of 530 nm. Totally, 90% and 50% inhibitory concentrations were taken as the relative MIC values [20, 21].Interaction of fluconazole/clotrimazole
The interaction between FLU and CLT was determined on the basis of the fractional inhibitory concentration (FIC) index. The FIC index was defined as follows: [(MIC of drug A in combination/MIC of drug A alone)] + [(MIC of drug B in combination/MIC of drug B alone)]. The interaction was defined as synergistic if the FIC index was ≤0.5, partial synergy when the FIC> 0.5 but < 1.0, additive if the FIC = 1.0, indifferent when the FIC index > 1.0 but < 4.0, and antagonistic if the FIC ≥ 4.0 [22].
Time-killing test
In order to further confirm the effect of FLU and CLT alone and in combination, C. albicans ATCC 14053 and CI-5- isolate were used to determine the time-killing curves. Initial inoculum at a density of 1×106 cells/ml were treated with different concentrations (2×MIC and 1×MIC) of FLU and CLT alone and in combination. The number of viable cells was determined by colony counting at 0, 2, 4, 6, 8, 10, 12, 24, and 48 h after exposure to antifungal agents at 35˚C. The interaction between FLU and CLT tested by time-kill methods (at 24 to 48 h) were determined as follows: synergy, ≥2 log10 decrease in CFU/ml compared to the most active agent; additive effect <2 but >1 log10 decrease in CFU/ml compared to the most active agent; indifference, <2 but >1 log10 increase in CFU/ml compared to the least active agent; and antagonism, a ≥2 log10 increase in CFU/ml compare to the least active agent [23, 24].
Combination of fluconazole/clotrimazole on C. albicans hyphae
The hyphae formation assay was performed in U-bottomed 96-well polystyrene microtiter plate as described previously [25]. Briefly, 100 μl of the C. albicans ATCC 14053 cell suspension (1×106 cells/ml) was dispensed into the wells of microtiter plates. Different concentrations (2×MIC, 1×MIC, ½×MIC and ¼×MIC) of FLU and CLT alone and in combination were added (100 μl per well) to each well and incubated at 35◦C for 90 min without agitation. The microtiter plate was incubated at 35◦C for 16 h with gentle shaking. The metabolic activity of the C. albicans hyphae was determined quantitatively using colorimetric XTT [2,3-bis (2-methoxy-4-nitro-5 sulfophenyl)-5-[(phenylamino) carbonyl 2]-H-tetrazolium hydroxide] reduction and crystal violet assays.
XTT reduction assay of C. albicans hyphae
At the end of appropriate incubation, the supernatant was aspirated from the wells and washed 3-times with sterile phosphate-buffered saline. The C. albicans cell viability was determined using colorimetric XTT reduction assay [25]. Sterilized XTT/menadione (Sigma-Aldrich) solution transferred to each well containing prewashed hypha and incubated in the dark at 37◦C for 5 h. After the incubation, colorimetric change in the XTT reduction was measured in a microtiter microplate reader at 490 nm.
Crystal violet assay of C. albicans hyphae
The prewashed C. albicans hyphae was fixed with methanol and air dried. Then, C. albicans hyphae was stained with crystal violet solution (Sigma-Aldrich) for 20 min. Hundred μl of acetic acid 33% (Sigma-Aldrich) was distributed into the wells containing prewashed hyphae. The optical density was measured at 590 nm with Stat Fax 303 Reader (Awareness Technology) [25-27].
Scanning electron microscopy of C. albicans hyphae
Standard cell suspension of C. albicans was diluted (1×106 cells/ml) in RPMI-1640 medium supplemented with combination of FLU/CLT at MIC concentration on ThermanoxTM plastic coverslips (Nunc, Denmark) in 6-well cell culture plates (Nunc). The cell culture plates were incubated at 35◦C for 90 min (without agitation) and incubated again at 35◦ C for 16 h with gentle shaking. Thereafter, the prewashed hyphae was fixed in 2% (v/v) glutaraldehyde in phosphate-buffered saline (PH 7.2). Samples were washed with sodium cacodylate buffer and placed in 1% osmium tetroxide for 2 h at 4◦ C.
Samples were subsequently washed with sodium cacodylate buffer, dehydrated in a series of ascending ethanol solutions, put into critical point dryer and then stuck onto the stub. The specimens were coated with gold and viewed with a Philips XL30 (ESEM, UK) scanning electron microscope [25].
qRT-PCR analysis of C. albicans hypha-specific gene
C. albicans hyphae was performed in the presence of standard cell suspension of C. albicans and different concentrations (2×MIC and 1×MIC) of FLU and CLT alone and in combination in 6-well cell culture plates (Nunc).
Total RNA was extracted from C. albicans hyphae using RNeasy Mini Kit (Qiagen, Hilden, Germany) and 0.5 µg of total RNA was reverse transcribed with Moloney-Murine Leukemia Virus (MMLV) reverse transcriptase and random hexamers (Fermentas, USA). Resultant cDNAs were amplified by real time polymerase chain reaction (RT-PCR) using hypha specific and housekeeping internal control beta-actin gene primers (Table 1). Real time PCR reactions were run with ™SYBR Green qPCR Master Mix (Fermentas, USA) on Bio-Rad MiniOpticonTM system (USA). Relative expression was quantified by the Pfaffl method [23, 25].
The Research Ethics Committees of our institute (Ethical code 1213469) approved the study. The study protocol conformed to the ethical guidelines of the 2013 Declaration of Helsinki. Informed consent was obtained from patients.
Statistical analysis
Data were expressed as mean values of the biological replicates±standard deviations. One-way analysis of variance was applied to test the differences between treated and control groups. Tukey's HSD test was performed for a multiple comparison and statistical significance was tested at the p<0.05 levels. Statistical analyses were performed using the software SPSS 21.0 for windows (SPSS Inc. Chicago, IL, USA).
Results
Table 2 illustrates the susceptibility of clinical isolates of C. albicans and reference strains of C. albicans ATCC 14053 to FLU alone and in combination with CLT. The isolates were considered resistant at the MIC value ≥8.0 μg/ml for fluconazole, and ≥1.0 μg/ml for CLT as established by the CLSI M27-A3. As presented in table 2, all clinical isolates of C. albicans were resistant to FLU (MIC range 12.30-18.90 μg/ml), and CLT (MIC range 1.25-2.50 μg/ml).
The combined effects of the FLU and CLT against clinical isolates of C. albicans and reference strains of C. albicans ATCC 14053 are shown in table 3. A comparison between these values showed that the combination of FLU and CLT can be decreased from its MIC value of FLU and CLT by 6.79- to 61.50-fold and 1.04- to 6.25-fold, respectively. FLU showed synergistic (70%), partial synergistic (20%) and indifferent (10%) interaction with CLT against clinical isolates of C. albicans.The synergism of FLU/CLT against C. albicans ATCC 14053 and CI-5 isolate were confirmed by time-killing curves (Fig. 1). The combination of FLU/CLT at 2×MIC and 1×MIC concentrations against C. albicans ATCC 14053 revealed a 4.06, 4.10, 3.16 and 3.60 log10 CFU /ml decrease compared with FLU alone at 24 and 48 h, respectively. The results indicated that FLU and CLT action alone have little antifungal effect throughout 48 h in CI-5- isolate of C. albicans, but the combination yielded 2 × MIC and 1 × MIC a 6.03, 4.80, 7.02 and 5.95 log10 CFU/ml decrease compared with fluconazole alone at 24 and 48 h, respectively.
Table 1. Primers for analysis of hyphae specific gene by RT-PCR
| Primer | Orientation | Sequence | Reference |
| HWP1 | Forward | 5´ TCAGTTCCACTCATGCAACCA 3´ | [28] |
| Reverse | 5´ AGCACCGAAAGTCAATCTCATGT 3´ | ||
| Beta-actin | Forward | 5´ GAGTTGCTCCAGAAGAACATCCAG 3´ | [29] |
| Reverse | 5´ TGAGTAACACCATCACCAGAATCC 3´ |
Table 2. Susceptibility of clinical isolates of C. albicans in terms of MIC (μg/ml) to FLU and CLT
| Isolates/ Antifungal agents | FLU | CLT | ||
| MIC90 | MIC50 | MIC90 | MIC50 | |
| C. albicans ATCC 14053 (Control) | 3.50±0.10 | 0.40±0.02 | 0.90±0.01 | 0.45±0.20 |
| CI- 1 | 12.30±0.30 | 6.70±0.02 | 1.25±0.10 | 0.50±0.10 |
| CI- 2 | 16.30±0.01 | 7.90±0.10 | 2.50±0.05 | 0.50±0.03 |
| CI- 3 | 16.50±0.03 | 8.80±0.10 | 2.50±0.04 | 0.50±0.01 |
| CI- 4 | 15.50±0.05 | 6.80±0.03 | 2.50±0.06 | 0.25±0.09 |
| CI- 5 | 16.50±0.04 | 3.70±0.03 | 2.50±0.04 | 0.25±0.02 |
| CI- 6 | 12.50±0.30 | 3.90±0.04 | 2.50±0.05 | 0.50±0.09 |
| CI- 7 | 13.50±0.04 | 6.60±0.10 | 1.70±0.09 | 0.25±0.08 |
| CI- 8 | 13.50±0.05 | 6.50±0.10 | 1.50±0.02 | 0.25±0.09 |
| CI- 9 | 18.50±0.10 | 6.60±0.09 | 1.50±0.08 | 0.25±0.02 |
| CI- 10 | 18.90±0.02 | 6.90±0.04 | 1.50±0.04 | 0.25±0.05 |
Table 3. Interaction of FLU and CLT against clinical isolates of C. albicans
| Isolates/ Antifungal agents | FLU/CLT | ||||
| MIC90 | MIC50 | FIC90 | FIC50 | Outcome | |
| C. albicans ATCC 14053 (Control) | 0.09±0.05 | 0.05±0.01 | 0.13 | 0.24 | Synergy |
| CI- 1 | 0.20±0.01 | 0.05±0.04 | 0.18 | 0.11 | Synergy |
| CI- 2 | 2.40±0.02 | 0.90±0.05 | 1.11 | 1.91 | Indifferent |
| CI- 3 | 0.90±0.04 | 0.06±0.04 | 0.42 | 0.13 | Synergy |
| CI- 4 | 0.90±0.05 | 0.07±0.09 | 0.42 | 0.29 | Synergy |
| CI- 5 | 1.50±0.01 | 0.19±0.05 | 0.69 | 0.81 | Partial Synergy |
| CI- 6 | 1.40±0.02 | 0.25±0.01 | 0.67 | 0.56 | Partial Synergy |
| CI- 7 | 0.50±0.01 | 0.09±0.09 | 0.33 | 0.37 | Synergy |
| CI- 8 | 0.50±0.05 | 0.09±0.02 | 0.37 | 0.37 | Synergy |
| CI- 9 | 0.50±0.01 | 0.08±0.01 | 0.36 | 0.33 | Synergy |
| CI- 10 | 0.50±0.09 | 0.07±0.00 | 0.36 | 0.29 | Synergy |
Table 4 exhibits the significant results to reduce hyphae after treatment with antifungal agents using XTT and CV assays for FLU and CLT alone and in combination in all concentrations based on MIC (p=0.001). XTT and CV assays for hyphae quantification also indicated C. albicans ATCC 14053 hyphae treated with combination of FLU and CLT being more significant than FLU and CLT alone (p=0.001).
Table 4. Results of XTT and CV assays against C. albicans ATCC 14053 hyphae treated with FLU and CLT alone and in combination in different concentrations based on MIC
| Concentration of antifungal agents | Means absorbance at 490 nm±SD using XTT assay | Means absorbance at 590 nm±SD using CV assay | ||||
| FLU | CLT | FLU/CLT | FLU | CLT | FLU/CLT | |
| 2 × MIC | 0.13 ± 0.01 | 0.15 ± 0.06 | 0.10 ± 0.03 | 1.38 ± 0.01 | 1.44 ± 0.08 | 1.09 ± 0.03 |
| 1 × MIC | 0.15 ± 0.01 | 0.18 ± 0.02 | 0.11 ± 0.02 | 1.44 ± 0.04 | 1.48 ± 0.09 | 1.11 ± 0.05 |
| ½ × MIC | 0.16 ± 0.02 | 0.19 ± 0.04 | 0.12 ± 0.02 | 1.48 ± 0.02 | 1.49 ± 0.07 | 1.19 ± 0.06 |
| ¼ × MIC | 0.18 ± 0.01 | 0.21 ± 0.01 | 0.13 ± 0.02 | 1.55 ± 0.04 | 1.61 ± 0.09 | 1.22 ± 0.05 |
| Untreated control | 0.43 ± 0.02 | 0.43 ± 0.03 | 0.43 ± 0.04 | 2.11 ± 0.01 | 2.11 ± 0.01 | 2.11 ± 0.01 |
The combination of FLU and CLT against C. albicans ATCC 14053 hyphae was visually verified by scanning electron microscopy (SEM) (Fig. 2). The growing hyphae was composed by yeast cells and hyphae form in untreated control group. The FLU and CLT combination-treated hyphae reduced in number and density of cells and also completely destroyed C. albicans hyphae.
In qRT-PCR, a variable Ct value of HWP1 gene in C. albicans ATCC 14053 hyphae treated with FLU and CLT alone and in combination revealed their gene expression levels being affected by the treatments (Fig. 3). The mean Ct values revealed that the combination of FLU/CLT induces a significant decrease in expression level of the HWP1 gene by 2.86- and 2.33-fold at 2× MIC and 1× MIC concentrations, respectively (p=0.002). Furthermore, the expression of HWP1 gene in C. albicans ATCC 14053 hyphae treated with FLU and CLT FLU and CLT alone showed a variable level of expression compared to the untreated control.


Fig. 1. Time-killing curves of FLU and CLT alone and in combination against C. albicans ATCC 14053 and CI-5 isolate at 0, 2, 4, 6, 8, 10, 12, 24, and 48 h. Data is represented as mean±SD.

Fig. 2. Scanning electron microscopy of C. albicans ATCC 14053 hyphae. (A) Untreated control, the morphology is composed of yeast cells and hyphae form. (B) Treated with combination of FLU and CLT at MIC concentration, complete destruction of hyphae form and reduced yeast cells were observed. Magnification 1250×.



Fig 3. Whisker box plots of HWP1 gene expression at different concentrations of FLU and CLT alone and in combination based on MIC in C. albicans ATCC 14053 hyphae. The boxes represent the mean Ct values and the line inside the box represents median values. The bars across the boxes show locations of the minimum and maximum Ct values. Whiskers in the plot represent 95% confidence intervals. Data represent mean values±SD.
Discussion
Therapeutic strategies for fungal infections are limited by the small number of available antifungal drugs so that there is a need for efficient methods to identify novel drug targets and develop effective drug combinations [23, 30]. The switch between yeast and hyphae is a major role in the pathogenesis of C. albicans that could serve as target for treatment of infections. Several studies have proven useful in achieving that goal [23, 30-32]. In the current report, we demonstrate the combination of FLU/CLT on C. albicans and uncover their effective combinations on hyphae formation.
A synergistic effect was observed when FLU combined with CLT in clinical isolates proved resistant to FLU and CLT. FLU and CLT alone and in combination inhibited hyphal formation; they inhibited cell proliferation at higher concentrations. Combination of FLU/CLT is an extensively investigated drug combination that was shown to inhibit Candida cells [19, 33]. In an attempt to investigate the effectiveness combination of FLU/CLT on C. albicans hyphae, the growing hyphae was verified by XTT and CV assays and SEM. The combination of FLU and CLT destroyed hyphae and yeast form, suggesting their effective combinations on hyphae formation. We then analysed the expression level of hyphae specific gene in the C. albicans hyphae. Of three treatments, combination of FLU/CLT decreased and more variable expression of hyphae specific gene; in addition, FLU and CLT alone demonstrated a variable level of expression thus suggesting a change in the expression of HWP1 gene involved in hyphae formation and pathogenesis. We confirmed that hyphae is a target of the combination of FLU/CLT by showing inhibition of cell proliferation and hyphae formation and molecular analysis of hyphae specific gene in C. albicans.
Microbial analysis has already indicated that azole antifungal drugs are effective on C. albicans hyphae formation [23, 31, 34, 35]. Likewise, a role for HWP1 in C. albicans hyphal growth had been demonstrated by adhesins to host epithelial cell receptors [2, 3, 7, 8]. Our study provides further support for this interaction by linking pharmacological inhibition of hyphae formation. The inhibition of hyphae formation was accompanied by a reduction in hyphae specific gene expression. Khodavandi et al. exhibited that combination of FLU/ amphotericin B reduced yeast-hyphal transition and biomass and metabolic activity of the hypha in C. albicans [23]. Wakabayashi et al. demonstrated that triazole antifungal agents combined with triazoles lactoferrin-related compounds inhibit the growth of hyphae, an important form of pathogenesis in azole-resistant C. albicans strains [30]. The synergistic antifungal effect of licofelone in combination with fluconazole against C. albicans strongly reduced hyphal formation. The expression of RAS/cAMP/PKA pathway related genes (RAS1, CYR1, TPK2), and biofilm formation related genes including biofilm and cell wall regulator 1 (BCR1), HWP1, agglutinin-like sequence (ALS) 1 and ALS3 genes were down-regulated with the presence of licofelone combined with fluconazole [31]. FLU in combination with CLT revealed significant synergistic effects against C. tropicalis with FIC indices ranging from 0.011 to 0.43. In addition, the combination of FLU with CLT could trigger a down-regulation of C. tropicalis agglutinin-like sequence (ALS1 and ALS3), lipase (LIP1 and LIP4), and secreted aspartyl protease (SAP2 and SAP4) genes, respectively [36].
Conclusion
Our intention in assaying the effectiveness combination of FLU/CLT on C. albicans was to investigate the hyphae formation and transcriptional responses. Our results suggest that the HWP1 might constitute useful targets for the combination of FLU and CLT. This raises the prospect of identifying molecular mechanisms of combination of FLU/CLT as potential inhibitors of C. albicans virulence. However, this study demonstrated that the antifungal effect of FLU in combination with CLT could be critical in developing therapeutic strategies against C. albicans.
Conflict of interest
The authors declared no conflict of interest.
Acknowledgement
This research was supported by Islamic Azad University of Yasooj, Yasooj, Iran.
References
- Whittington A, Gow NAR, Hube B. From commensal to pathogen: Candida albicans. In: Kurzai O, editor. Human fungal pathogens. The mycota XII. Berlin: Springer-Verlag; 2014. pp. 3-18.
- Moyes DL, Richardson JP, Naglik JR. Candida albicans-epithelial interactions and pathogenicity mechanisms: scratching the surface. Virulence 2015; 6(4): 338-46.
- da Silva Dantas A, Lee KK, Raziunaite I, Schaefer K, Wagener J, Yadav B1 Gow NA. Cell biology of Candida albicans-host interactions. Curr Opin Microbiol. 2016; 34(2): 111-18.
- Jabra-Rizk MA, Kong EF, Tsui C, Nguyen MH, Clancy CJ, Fidel PL, et al. Candida albicans pathogenesis: fitting within the host-microbe damage response framework. Infect Immun. 2016; 84(10): 2724-739.
- Sharifynia S, Badali H, Sharifi Sorkherizi M, Shidfar MR, Hadian A, Shahrokhi S, et al. In vitro antifungal susceptibility profiles of Candida albicans complex isolated from patients with respiratory infections. Acta Med Iran. 2016; 54(6): 376-81.
- Achkar JM, Fries BC. Candida infections of the genitourinary tract. Clin Microbiol Rev. 2010; 23(2): 253-73.
- Klis FM, Sosinska GJ, de Groot PWJ, Brul S. Covalently linked cell wall proteins of Candida albicans and their role in fitness and virulence. Fems Yeast Res. 2009; 9(7): 1013-1028.
- Desai JV. Candida albicans hyphae: from growth initiation to invasion. J Fungi (Basel). 2018; 4(1): 10.
- Sharkey LL, McNemar MD, Saporito Irwin SM, Sypherd PS, Fonzi WA. HWP1 functions in the morphological development of Candida albicans downstream of EFG1, TUP1, and RBF1. J Bacteriol. 1999; 181(17): 5273-279.
- Staab JF, Bahn YS, Tai CH, Cook PF, Sundstrom P. Expression of transglutaminase substrate activity on Candida albicans germ tubes through a coiled, disulfide-bonded N-terminal domain of Hwp1 requires C-terminal glycosylphosphatidylinositol modification. J Biol Chem. 2004; 279(39): 40737-40747.
- Kaur H, Chakrabarti A. Strategies to reduce mortality in adult and neonatal candidemia in developing countries. J Fungi (Basel). 2017; 3(3): 41.
- Bongomin F, Gago S, Oladele RO, Denning DW. Global and multi-national prevalence of fungal diseases-estimate precision. J Fungi (Basel). 2017; 3(4): 57.
- Lamoth F, Lockhart SR, Berkow EL, Calandra T. Changes in the epidemiological landscape of invasive candidiasis. J Antimicrob Chemother. 2018; 73(S1): 4-13.
- Pappas PG, Kauffman CA, Andes DR, Clancy CJ, Marr KA, Ostrosky-Zeichner L, et al. Clinical practice guideline for the management of candidiasis: 2016 update by the infectious diseases society of America. Clin Infect Dis. 2016; 62(4): 1-50.
- Pelletier R, Peter J, Antin C, Gonzalez C, Wood L, Walsh TJ. Emergence of resistance of Candida albicans to clotrimazole in human immunodeficiency virus-infected children: in vitro and clinical correlations. J Clin Microbiol. 2000; 38(4): 1563-568.
- Berkow EL, Lockhart SR. Fluconazole resistance in Candida species: a current perspective. Infect Drug Resist 2017; 10(1): 237-45.
- Belanger ES, Yang E, Forrest GN. Combination antifungal therapy: when, where, and why. Curr Clin Microbiol Rep. 2015; 2(2): 67-75.
- Campitelli M, Zeineddine N, Samaha G, Maslak S. Combination antifungal therapy: a review of current data. J Clin Med Res. 2017; 9(6): 451-456.
- Mendling W, Krauss C, Fladung B. A clinical multicenter study comparing efficacy and tolerability of topical combination therapy with clotrimazole (Canesten, two formats) with oral single dose fluconazole (Diflucan) in vulvovaginal mycoses. Mycoses. 2004; 47(3-4): 136-42.
- Alizadeh F, Khodavandi A, Zalakian S. Quantitation of ergosterol content and gene expression profile of ERG11 gene in fluconazole-resistant Candida albicans. Curr Med Mycol. 2017; 3(1): 13-19.
- Clinical and Laboratory Standards Institute (CLSI). Reference method for broth dilution antifungal susceptibility testing of yeasts; Approved standard M27-A3. Wayne PA, Clinical and Laboratory Standards Institute: 2008.
- Khodavandi A, Alizadeh F, Aala F, Sekawi Z, Chong PP. In vitro investigation of antifungal activity of allicin alone and in combination with azoles against Candida species. Mycopathologia 2010; 169(4): 287-95.
- Khodavandi A, Alizadeh F, Khezrian F. Inhibition of Candida albicans yeast– hyphal transition by combination of fluconazole with amphotericin B. Physiol Pharmacol. 2019; 23(1): 195-204.
- Mukherjee PK, Sheehan DJ, Hitchcock CA, Ghannoum MA. Combination treatment of invasive fungal infections. Clin Microbiol Rev. 2005; 18(1): 163-94.
- Khodavandi A, Harmal NS, Alizadeh F, Scully OJ, Sidik SM, Othman F, et al. Comparison between allicin and fluconazole in Candida albicans biofilm inhibition and in suppression of HWP1 gene expression. Phytomedicine 2011; 19(1): 56-63.
- Peeters E, Nelis HJ, Coenye T. Comparison of multiple methods for quantification of microbial biofilms grown in microtiter plates. J Microbiol Methods 2008; 72(2): 157-165.
- Braga PC, Culici M, Alfieri M, Dal Sasso M. Thymol inhibits Candida albicans biofilm formation and mature biofilm. Int J Antimicrob Agents 2008; 31(5): 472-77.
- Uppuluri P, Chaturvedi AK, Lopez-Ribot JL. Design of a simple model of Candida albicans biofilms formed under conditions of flow: development, architecture, and drug resistance. Mycopathologia 2009; 168(3): 101-109.
- Lim CS, Wong WF, Rosli R, Ng KP, Seow HF, Chong PP. 2-dodecanol (decyl methyl carbinol) inhibits hyphal formation and SIR2 expression in C. albicans. J Basic Microbiol. 2009; 49(6): 579-83.
- Wakabayashi H, Abe S, Teraguchi S, Hayasawa H, Yamaguchi H. Inhibition of hyphal growth of azole-resistant strains of Candida albicans by triazole antifungal agents in the presence of lactoferrin-related compounds. Antimicrob Agents Chemother. 1998; 42(7): 1587-591.
- Liu X, Li T, Wang D, Yang Y, Sun W, Liu J, et al. Synergistic antifungal effect of fluconazole combined with licofelone against resistant Candida albicans. Front Microbiol. 2017; 8(1): 2101.
- Moazeni M, Khoramizadeh MR, Teimoori Toolabi L, Noorbakhsh F, Rezaie S. The effect of EFG1 gene silencing on down-regulation of SAP5 gene, by use of RNAi technology. Acta Med Iran. 2014; 52(1): 9-14.
- Gharibi T, Ganjoo M, Kamali F, Ahmadi S, Pouladi S, Vahed parast H, et al. Comparison of combined use of fluconazole and clotrimazole with the sequential dose of fluconazole in the treatment of recurrent Candida vaginitis. Iran South Med J. 2009; 12(1): 34-39.
- Ha KC, White TC. Effects of azole antifungal drugs on the transition from yeast cells to hyphae in susceptible and resistant isolates of the pathogenic yeast Candida albicans. Antimicrob Agents Chemother. 1999; 43(4): 763-68.
- Calabrese EC, Castellano S, Santoriello M, Sgherri C, Quartacci MF, Calucci L, et al. Antifungal activity of azole compounds CPA18 and CPA109 against azole-susceptible and -resistant strains of Candida albicans. J Antimicrob Chemother. 2013; 68(5): 1111-119.
- Khodavandi A, Alizadeh F, Abdolahi M, Jahangiri M. Differential expression levels of agglutinin-like sequence, lipase, and secreted aspartyl protease genes in Candida tropicalis treated with fluconazole alone and in combination with clotrimazole. J Rep Pharma Sci. 2019; 8(1): 28-33.
Send email to the article author
| Rights and permissions | |
 |
This work is licensed under a Creative Commons Attribution-NonCommercial 4.0 International License. |