Helicobacter pylori (H. pylori) is a microaerophilic, gram-negative bacterium that is recognized to be one of the leading causes of gastroduodenal ulcers, gastritis, and gastric cancer [1, 2]. Previous studies about the presence of this bacterium in various specimens of the oral cavity such as dental plaque and saliva revealed that the oral cavity could be a considerable reservoir for H. pylori or as a potential route of transmission to other origins of body. The oral cavity can be the second origin of the homing of H. pylori that may result in recurrent gastric infection [3]. The reported prevalence of H. pylori infection in Asia is 11% to 90%, Africa is 48% to 90%, Central America is 51% to 90%, North America is 7% to 30%, and South America is 54% to 90%, and Europe is observed from 1.2% to 70.7% [4].
Epidemiologic studies conducted in Iran have reported the prevalence of H. pylori in the range of 82-92% [5, 6]. Since this bacterium is microaerophilic and requires special conditions for colonization, oral cavity, dental plaque, and periodontal plaques can be significant places for the storage of this bacterium [7]. Despite the impact of multiple genes on the virulence of bacteria, extensive studies could not specify a definitive relationship between the clinical consequences of an infection and a particular gene. In any case, there is a belief among researchers that virulence strains are more likely to cause gastrointestinal diseases, especially gastric ulcers [8-10]. The most critical pathogenicity factors of H. pylori include urease, flagellum, addict, cacto vaccine (vacA, cagA) and pathogenicity island (cagPAI) [11]. IceA gene and H. pylori have recently been recognized as a genetic indicator for the development of duodenal ulcer disease in the East [12]. Peek and colleagues introduced the gene. AceA has two iceA1, iceA2 alleles. The iceA1 allele codes for the homolog of the nlaIIIR endonuclease enzyme in N. lactamica, and iceA2 codes the protein with 59 amino acids and does not depend on iceA1. The role of iceA1 gene in human infection has not been determined yet [13-15]. The expression of one of these two depends on the prevalence of the disease and the type of disease. Studies have shown that the iceA1 allele is linked to a peptic ulcer in the Netherlands and the United States but not in countries such as Japan, Korea, and Colombia [12, 15]. Finally, this study aimed to find out the presence of this bacterium in dental plaque in subjects without symptoms. Therefore, due to the high prevalence of gastrointestinal disorders such as non-ulcers and wounded indigestion, the probability of the bacteria storage in the oral cavity and the percentage of iceA gene is evaluated.
Materials and Methods
This study was conducted during 2 years at the Genetics Laboratory of Sari Islamic Azad University. One hundred samples of plaque were taken from Bojnourd city. As a dental plaque was drawn with sterilized swabs, it was pulled into sterilized physiology serum. At the end of each day, the samples were transferred to -70 °C until the DNA could be extracted. Then, by using the phenol-chloroform protocol, DNA was extracted and subjected to spectrophotometry for quantification. DNA extracts were used to perform the polymerase chain reaction (PCR) technique to detect H. pylori by primer sets C97-20 [5′-GGCTATGACGGGTATCCGGC3′]; positions [260-279] and H3A-20 [5′-GCCGTGCAGCACCTGTTTTC-3′]; anneals to positions 1007 to 1026 of H. pylori 16S rRNA that amplify a fragment with the size of 746 bp and also iceA gene representing H. pylori strains by F- ATTACTGACGCTGATTGTGC and R- CTGGAGAGACTAAGCCCTCC primers that can amplify a DNA fragment with the size of 250 bp. We used the H.pylori positive sample as the positive control. Eppendorf thermocycler was used for DNA amplification (Roche Co., Germany). Amplification for 16s rRNA was performed in a final volume of 25 µl containing 1 µl of MgCl2, 0.3 µl of dNTP, 0.3 µl of each primer, 0.2 µl of Taq polymerase, 2.5 µl PCR buffer, 1.5 µl of template DNA and 18.9 µl double-distilled water. DNA amplification for iceA gene was performed in a final volume of 25 µl containing 1 µl of MgCl2, 0.3 µl of dNTP, 0.4 µl of each primer, 0.2 µl of Taq polymerase, 2.5 µl PCR buffer, 1.5 µl of template DNA and 18.7 µl double distilled water. The amplification for 16s rRNA was performed with an initial denaturation at 94°C for 4 min, followed by 30 cycles of denaturation at 94°C for 60 s, annealing at 57°C for 45 s and extension at 72°C for 60 s with a final extension at 72°C for 10 min. The amplification for iceA gene was performed with an initial denaturation at 95°C for 4 min, followed by 30 cycles of denaturation at 95°C for 60 s, annealing at 56°C for 45 s and extension at 72°C for 60 s with a final extension at 72°C for 10 min. The PCR products were separated on 1% (w/v) agarose gel with 0.5 mg/ml of loading dye and were analyzed by gel electrophoresis. This study was approved by the Ethics Committee of Islamic Azad University, Sari, Iran.
Statistical analysis
Statistical Package for the Social Sciences software (SPSS Inc No. 22) was used for data analysis.
Result
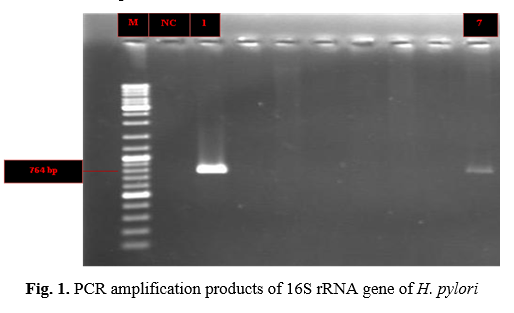
In this study, 98 samples of tooth plaque in Bojnourd city were taken, 2% of them were positive for H. pylori (Table 1), and both positive H. pylori were positive for iceA gene. Fig. 1. shows PCR amplification products of 16S rRNA gene of H. pylori. PCR amplification products of iceA gene of H. pylori is presented in Fig. 2.

Table 1. The results of the frequency of
H. pylori
| Sample |
Frequency |
Frequency (%) |
Cumulative frequency (%) |
| Positive |
2 |
2 |
2 |
| Negative |
98 |
98 |
98 |
| Total |
100 |
100 |
100 |
Discussion
H. pylori infection is one of the important and common gastric infections all around the world. Recently, these microorganisms have been detected in dental plaques, where the prevalence of these bacteria was supposed to be very low. The presence of bacteria appears to be different in various sites within the oral cavity [16]. The present study identified DNA from H. pylori in 2% of clinical dental plaque specimens. Indeed, H. pylori infection is affected by public health and low level of economic welfare, so that the prevalence of H. pylori in Iran and Brazil is higher than 80% [17,18]. Besides, 72% of patients with the gastric infection have H. pylori in their dental plaque in Brazil. Even though this result was in contrast with our result due to having a 2% presence of H pylori, the amount of dental plaque in the oral cavity has been shown by some researchers to have a direct influence on the rate of H. pylori detection in study samples. This implies that improving oral hygiene by regular tooth brushing would improve the oral hygiene status and result in clearance of the bacterium from the mouth. The present investigation found no significant relationship between the frequency of brushing and the presence of H. pylori in the understudied dental plaque samples. Nearly 2% of the individuals were positive for the bacterium in their dental plaque samples. Assumpcao et al. (2010) conducted a study called H. pylori in the teeth and stomach plaque of northern Brazilian patients. In this study, a cavity plasticized sample and a gastric biopsy specimen were used for histological examination, rapid urease test, and PCR to detect the presence of cagA and vacA gene. The results of this study indicated that the detection of H. pylori from tooth plaque and gastric biopsy specimens was done mostly by PCR in comparison with the histological examination and urease test. The DNA of H. pylori was observed in 96% of the gastric mucosal specimen and 72% of the dental plaque samples. Sixty-three samples (89%) of 71 plaque duplicates that were positive in H. pylori have vacA and cagA genes [19]. In 2008, Shirvani et al. conducted a study on H. pylori strains isolated from people with gastroduodenal diseases in Babol city.
Among 50 patients with gastroduodenal gastric biopsy, CagA and iceA (iceA2 and iceA1 alleles) were examined for the presence of these genes. After cultivating and extracting DNA using specific primers for CagA, iceA, (iceA2 and iceA1) genes, PCR was tested. The results of this study showed that the prevalence of CagA was higher in H. pylori strains, and the iceA allele was also recognized as the dominant allele in this region [20]. In the present study, two positive iceA samples were not included in any of the iceA2 and iceA1 alleles that appear to be new alleles of this gene. In this study, the gene of iceA was investigated, just like the research. According to previous studies, 100 samples were collected, and 100 samples of these bacteria have been detected in 2% of the samples. Besides, both of the H. pylori had an Ice A gene, which means 100% of H. pylori isolated from the dental plaques were positive for iceA gene.
Conclusion
Although the frequency of bacterial detection in dental plaque samples was low, the presence of iceA gene in all Helicobacter samples was positive. However, due to the high genetic diversity among H. pylori strains in different societies, further research on the role of this gene in the development of digestive diseases seems necessary.
Conflict of Interest
The authors declared no conflict of interests.
Acknowledgment
The authors have not expressed any acknowledgment.
Reference
- Dunn BE, Cohen H, Blaser MJ. H. pylori. Clin Microbiol Rev. 1997; 10(1): 720-41.
- Taylor DN, Blaser MJ. The epidemiology of H. pylori infection. Epidemiol Rev. 1991; 13(1): 42-59.
- Miyabayashi H, Furihata K, Shimizu T, Ueno I, Akamatsu T. Influence of oral H. pylori on the success of eradication therapy against gastric H. pylori. Helicobacter 2000; 5(5): 30-7.
- Hunt RH, Xiao SD, Megraud F, Leon-Barua R, Bazzoli F, van der Merwe S, et al. H. pylori in developing countries. World Gastroenterology Organisation Global Guidelines 2010; p. 4-5.
- Fakheri H, Merat S, Hosseini V, Malekzadeh R. Low dose furazolidone in triple and quadruple regimens for H. pylori eradication. Aliment Pharmacol Ther. 2004; 19(2): 89-93.
- Alborzi A, Soltani J, Pourabbas B, Oboodi B, Haghighat M, Hayati M, et al. Prevalence of H. pylori infection in children (south of Iran). Diagn Microbiol Infect Dis. 2006; 54: 259-61.
- Kawasaki ES. Sample preparation from blood, cells, and other fluids in PCR protocols: A Guide to Mmethods and Aapplications, 1st ed. Google books, 1990; p. 146-52.
- Salehi Z, Jelodar M, Rassa M, Ahaki M, Mollasalehi H, Mashayekhi F. H. pylori cagA status and peptic ulcer disease in Iran. Digest Dis Sci. 2009; 54(3): 608-13.
- Oliveira M, Costa A, Costa A, Henriques L, Suriano G, Atherton J, et al. H. pylori induces gastric epithelial cell invasion in a c- Met and type IV secretion system-dependent manner. J Biologic Chem. 2006; 281(46): 348-88.
- Nguyen L, Uchida T, Tsukamoto Y, Kuroda A, Okimoto T, Kodama M, et al. H. pylori dupA gene is not associated with clinical outcomes in the Japanese population. Clin Microbiol Infect. 2010; 16(8): 1264-269.
- Kundu P, Mukhopadhyay AK, Patra R, Fauchere JL. Cag pathogenicity island-independent up-regulation of matrix metalloproteinases- 9 and -2 secretion and expression in mice by H. pylori infection. J Biol Chem. 2006; 281(45): 34651-4662.
- Siavoshi F, Shokouhfard M, Malekzadeh R, Dinparast Jadid N, Massarrat S, Emrani A. Genotype Variation in H. Pylori Isolates from Iranian Patients by RAPD-PCR. J Govaresh. 2004(9): 1-10.
- Montecucco C, Rappuoli R. Living dangerously: how Heli-cobacter pylori survives in the human stomach. Nat Rev Mol Cell Biol. 2001; 2(2): 457-66.
- Achtman M, Suerbaum S. H. pylori: molecular and cellular biology, United Kingdom: Wymondham, Horizon Scientific Press; 2001.
- Marshall BJ, Barrett LJ, Prakash C, McCallum, RW, Guerrant RL. Urea protects Helicobacter (Campylobacter) pylori from the bac-tericidal effect of acid. Gastroenterol. 1990; 99(2): 697-702.
- Kilmartin CM. Dental implications of H. pylori. J Can Dent Assoc. 2002; 68(4): 489-93.
- Souto FJ, Fontes CJ, Rocha GA, de Oliveira AM, Mendes EN, Queiroz DM. Prevalence of H. pylori infection in a rural area of the state of Mato Grosso, Brazil. Mem Inst Oswaldo Cruz. 1998;93: 171-74.
- Mitchell A, Silva TM, Barrett LJ, Lima AA, Guerrant RL. Age-specific H. pylori seropositivity rates of children in an impoverished urban area of northeast Brazil. J Clin Microbiol. 2003; 41(5):1326-328
- Assumpcao MB, Martins LC, Peura D. H. pylori in dental plaque and stomach of patients from North Brazil. World J Gastroenterol. 2010; 16(24): 3033-3039.
- Shirvani R, Rajab Nia, Tohidi F, Asmar M, Taheri H. Frequency of cagA and iceA gene in H. pylori isolates isolated from individuals with gastroduodenal diseases in Babol city. Babol Med J. 2012; 10(3): 53-46.