Wed, Feb 4, 2026
[Archive]
Volume 9, Issue 2 (May 2022)
IJML 2022, 9(2): 150-155 |
Back to browse issues page
Download citation:
BibTeX | RIS | EndNote | Medlars | ProCite | Reference Manager | RefWorks
Send citation to:



BibTeX | RIS | EndNote | Medlars | ProCite | Reference Manager | RefWorks
Send citation to:
Shakeri Fini Z, Kheirmand Parizi P, Esmaeili S, Samadi-Khouzani A, Hosseini S, Jafarpour M. An Investigation of the Relationship Between HLA-CW06 and Psoriasis in Iranian Patients Using Sequence-specific Primers. IJML 2022; 9 (2) :150-155
URL: http://ijml.ssu.ac.ir/article-1-453-en.html
URL: http://ijml.ssu.ac.ir/article-1-453-en.html
Zahra Shakeri Fini 
 , Payam Kheirmand Parizi
, Payam Kheirmand Parizi 
 , Seyed-Alireza Esmaeili
, Seyed-Alireza Esmaeili 
 , Abbas Samadi-Khouzani
, Abbas Samadi-Khouzani 
 , Sara Hosseini
, Sara Hosseini 
 , Mostafa Jafarpour *
, Mostafa Jafarpour * 


 , Payam Kheirmand Parizi
, Payam Kheirmand Parizi 
 , Seyed-Alireza Esmaeili
, Seyed-Alireza Esmaeili 
 , Abbas Samadi-Khouzani
, Abbas Samadi-Khouzani 
 , Sara Hosseini
, Sara Hosseini 
 , Mostafa Jafarpour *
, Mostafa Jafarpour * 

Department of Microbiology, Faculty of Science, Islamic Azad University, Tonekabon Branch, Tonekabon, Iran
Full-Text [PDF 322 kb]
(557 Downloads)
| Abstract (HTML) (1395 Views)
References
Full-Text: (501 Views)
Introduction
Psoriasis is a chronic inflammatory disease with a worldwide distribution, affecting 0.44% to 2.88% worldwide of the general population. The prevalence of psoriasis has increased, and it is the second-largest contributor to skin-related disability [1-4]. It is a heterogeneous disorder with distinguished but overlapping phenotypes. This difference may be due to racial factors. Psoriasis influences various organs past the skin and has been related to metabolic dysfunction and cardiovascular sicknesses [5-8]. The pathogenesis of psoriasis is not fully known, but genetic history’s impact is a consensus issue [9, 10]. The human leukocyte antigen (HLA) located on chromosome 6p21 is notable for carrying the main hereditary elements in the vulnerability to psoriasis, whereas the critical segment seems to be a 300-kb distance at the centromeric end of HLA class I(6p21.33), named PSORS1[11-14]. The association of HLA and diseases is accepted. Many population studies offered to suggest proof of association of HLAs in additional than forty diseases [15, 16].
This study aimed to survey the HLA-CW6 distribution in Iranian psoriatic patients. HLA-Cw6 is known to be quite possibly the most emphatically related psoriasis susceptibility alleles. It is widely recognized that HLA-Cw6 is related to phenotypic highlights, for example, beginning stage psoriasis and guttate lesions[17]. Type I manifests early in life (onset < 40 years) and is possibly related to family history; type II onsets are later in life (onset > 40 years) and is more likely to be sporadic [18]. Stress, obesity, and streptococcal pharyngitis are generally seen in HLA-Cw6-positive patients [19, 20].
Materials and Methods
Research participants
The present case-control study was conducted to evaluate the relation between HLA-CW6 and psoriasis in a population in Iran. The target population is in northern Iran. This study was performed on 30 samples of patients with psoriasis. For the control group, 30 healthy individuals without any disease manifestation were collected, and age, sex, and geographical distribution were adjusted to the patient group. The sample size was calculated using the power calculator for case-control genetic association studies [21]. People in the control group should not have a family relationship with patients. At last, blood sampling was performed after obtaining their informed consent.
DNA extraction and genotyping
Peripheral blood samples of patient and control groups were collected in tubes with ethylenediaminetetraacetic acid as an anticoagulant. According to the manufacturer’s instructions, DNAs were then extricated from whole-blood samples by a DNA extraction Kit (GENET BIO, Chungnam, Korea). DNA quality was checked by the NanoDrop micro-volume spectrophotometer (Thermo Scientific NanoDrop Products) and through the agarose gel electrophoresis. The classic sequence-specific primers (SSP) HLA-Cw06 standard kit (Bio-Rad, United States) was used in the study. The applied primers for the polymerase chain reaction (PCR) amplification were as follows:
F: 5’-TACTACAACCAGAGCGAGGA-3'/
R: 5’-GGTCGCAGCCATACATCCA-3'–control primer F: 5'-TGCCAAGTGGAGCAC CCAA-3'/
R: 5'-GCATCTTGCTCTGTGCAGAT-3’.
PCR was performed on an Eppendorf thermal cycler (Eppendorf, Hamburg, Germany) under the following conditions: 95 °C for 5 min followed by 30 cycles of 96 °C for 25s, 65 °C for 50 s, 72 °C for 30 s, and a final extension of 72 °C for 1.5 min. In order to prevent the extraction buffer from any damage, the DNA extraction buffer should generally be stored at 4 °C. On top of that, for long-term DNA storage and to prevent any enzymatic and physical damage to the extraction supply, DNA would be kept at -20 °C.
Qualitative and quantitative analysis
This study aimed to survey the HLA-CW6 distribution in Iranian psoriatic patients. HLA-Cw6 is known to be quite possibly the most emphatically related psoriasis susceptibility alleles. It is widely recognized that HLA-Cw6 is related to phenotypic highlights, for example, beginning stage psoriasis and guttate lesions[17]. Type I manifests early in life (onset < 40 years) and is possibly related to family history; type II onsets are later in life (onset > 40 years) and is more likely to be sporadic [18]. Stress, obesity, and streptococcal pharyngitis are generally seen in HLA-Cw6-positive patients [19, 20].
Materials and Methods
Research participants
The present case-control study was conducted to evaluate the relation between HLA-CW6 and psoriasis in a population in Iran. The target population is in northern Iran. This study was performed on 30 samples of patients with psoriasis. For the control group, 30 healthy individuals without any disease manifestation were collected, and age, sex, and geographical distribution were adjusted to the patient group. The sample size was calculated using the power calculator for case-control genetic association studies [21]. People in the control group should not have a family relationship with patients. At last, blood sampling was performed after obtaining their informed consent.
DNA extraction and genotyping
Peripheral blood samples of patient and control groups were collected in tubes with ethylenediaminetetraacetic acid as an anticoagulant. According to the manufacturer’s instructions, DNAs were then extricated from whole-blood samples by a DNA extraction Kit (GENET BIO, Chungnam, Korea). DNA quality was checked by the NanoDrop micro-volume spectrophotometer (Thermo Scientific NanoDrop Products) and through the agarose gel electrophoresis. The classic sequence-specific primers (SSP) HLA-Cw06 standard kit (Bio-Rad, United States) was used in the study. The applied primers for the polymerase chain reaction (PCR) amplification were as follows:
F: 5’-TACTACAACCAGAGCGAGGA-3'/
R: 5’-GGTCGCAGCCATACATCCA-3'–control primer F: 5'-TGCCAAGTGGAGCAC CCAA-3'/
R: 5'-GCATCTTGCTCTGTGCAGAT-3’.
PCR was performed on an Eppendorf thermal cycler (Eppendorf, Hamburg, Germany) under the following conditions: 95 °C for 5 min followed by 30 cycles of 96 °C for 25s, 65 °C for 50 s, 72 °C for 30 s, and a final extension of 72 °C for 1.5 min. In order to prevent the extraction buffer from any damage, the DNA extraction buffer should generally be stored at 4 °C. On top of that, for long-term DNA storage and to prevent any enzymatic and physical damage to the extraction supply, DNA would be kept at -20 °C.
Qualitative and quantitative analysis
The accuracy of DNA extraction is so important that the extracted DNA is qualitatively and then quantitatively analyzed for molecular investig-ations. Electrophoresis and spectrophotometry were the techniques followed for qualitative and quantitative analysis.
Statistical analysis
Allele frequencies in the study group and the controls were evaluated by direct counting. Data were tested for the efficiency between the observed and expected genotype frequencies, and no deviations from the Hardy–Weinberg equilibrium were found. Associations of particular alleles with psoriasis were expressed as odds ratios (OR) with 95 % confidence intervals (CI) calculated according to Woolf’s formula [22].
Results
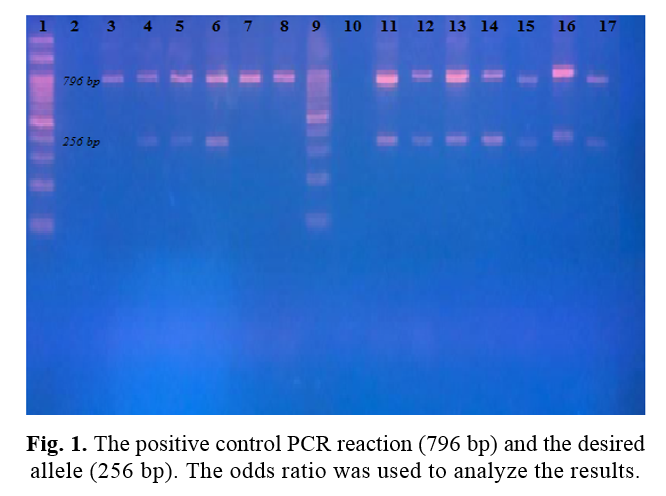
Existing case-control research studied a total of 60 volunteers. Among them, 30 patients with a history of psoriasis were classified as the case group. The results of PCR-SSP for the detection of HLA-CW06 showed positive amplification in 7 out of 30 psoriasis patients compared to 4 among 30 controls (Table 1). The electrophoresis image confirms the positive control PCR reaction (796 bp) and the desired allele (256 bp) (Fig. 1). According to the calculations, the OR was 1.97; due to the high number associated with psoriasis, we can say that the HLA allele affects this disease.
Discussion
Scientists estimate that about 10% of the world’s population has at least one gene that causes psoriasis, but the prevalence of the disease in the world is only 2 to 3%. In this case, the combination of genes that cause psoriasis and environmental factors contribute to the disease occurrence [17]. About one-third of people with psoriasis have a family history of the disease. Studies on identical twins showed that if one of the twins has psoriasis, there is a 70% chance that the other twins will also get the disease.
Allele frequencies in the study group and the controls were evaluated by direct counting. Data were tested for the efficiency between the observed and expected genotype frequencies, and no deviations from the Hardy–Weinberg equilibrium were found. Associations of particular alleles with psoriasis were expressed as odds ratios (OR) with 95 % confidence intervals (CI) calculated according to Woolf’s formula [22].
Results
Existing case-control research studied a total of 60 volunteers. Among them, 30 patients with a history of psoriasis were classified as the case group. The results of PCR-SSP for the detection of HLA-CW06 showed positive amplification in 7 out of 30 psoriasis patients compared to 4 among 30 controls (Table 1). The electrophoresis image confirms the positive control PCR reaction (796 bp) and the desired allele (256 bp) (Fig. 1). According to the calculations, the OR was 1.97; due to the high number associated with psoriasis, we can say that the HLA allele affects this disease.
Discussion
Scientists estimate that about 10% of the world’s population has at least one gene that causes psoriasis, but the prevalence of the disease in the world is only 2 to 3%. In this case, the combination of genes that cause psoriasis and environmental factors contribute to the disease occurrence [17]. About one-third of people with psoriasis have a family history of the disease. Studies on identical twins showed that if one of the twins has psoriasis, there is a 70% chance that the other twins will also get the disease.
Table 1. Comparison of the psoriasis types among HLA-Cw06-positive and HLA-Cw06-negative patients
| Absence of risk factor | Existence of risk factor |
| B=23 | A=7 | Patients |
| D=26 | C=4 | Controls |

On top of that, the probability for heterogeneous twins is 20%. These studies show the importance of genes’ impacts on psoriasis and the influence of environmental factors on the incidence and occurrence of psoriasis [23-26]. Psoriasis has a vital genetic component, so many genes are associated with the disease, but these genes causing the disease are still unknown. Today, researchers have identified many chromosomal loci associated with psoriasis. Most of the identified effective genes are related to the immune system, especially T cells and the major histocompatibility complex [27, 28]. In general, nine chromosomal sites associated with psoriasis have been identified, known as psoriasis susceptibility sites 1 to 9 or, for short, PSORS1 to PSORS9. These chromosomal sites are genes for the inflammatory pathways in patients with psoriasis; these genes are mutated. Other genes have also been discovered linked to the development of psoriasis and have been significantly altered in people with psoriasis. Some of these genes cause the expression of inflammatory proteins affecting cells in the immune system that are involved in causing psoriasis. Changes in some of these genes are also seen in other autoimmune diseases[29-33]. The most important effect of genes on psoriasis is related to the PSORS1 locus, responsible for 35 to 50% of hereditary psoriasis. PSORS1 is responsible for regulating immune-stimulating genes or producing skin proteins that are overproduced in psoriasis. The HLA-CW06 gene is one of the essential genes in the PSORS1 locus [34, 35]. The HLA-C* 06 allele is the genetic variation related to the phenotypic highlights of psoriasis, which have been seen more than once. It is associated mainly with the beginning stage of the illness, as affirmed by Szczerkowska-Dobosz et al. [36]. This study examined the interplay between the HLA-C allele and psoriasis in patients with PCR-SSP in the Iranian population. Given the different allelic frequencies associated with psoriasis in different parts of the world, it seems that other genetic and epigenetic factors are also involved in this disease. Nonetheless, broad approvals are needed to test the consensus of our theory among various populations around the world.
Conclusion
Our study confirmed the strong positive association of psoriasis with HLA-C*06, observed mainly in Caucasoid populations and in several other ethnically different groups.
Conflicts of Interest
The authors declare no conflicts of interest and financial support for the present original article.
Acknowledgments
We would like to thank the Department of Biology, Faculty of Basic Sciences, Shahrekord Branch, Islamic Azad University for their financial support.
Conclusion
Our study confirmed the strong positive association of psoriasis with HLA-C*06, observed mainly in Caucasoid populations and in several other ethnically different groups.
Conflicts of Interest
The authors declare no conflicts of interest and financial support for the present original article.
Acknowledgments
We would like to thank the Department of Biology, Faculty of Basic Sciences, Shahrekord Branch, Islamic Azad University for their financial support.
References
- Mehrmal S, Uppal P, Nedley N, Giesey RL, Delost GR. The global, regional, and national burden of psoriasis in 195 countries and territories, 1990 to 2017: A systematic analysis from the Global Burden of Disease Study 2017. J Am Acad Dermatol. 2021; 84(1): 46-52.
- Bhende S, Parwe S. Role of Nitya Virechana and Shaman Chikitsa in the management of Ekakushta with special respect to plaque psoriasis: A case study. J Indian Sys Med. 2020; 8(1): 57.
- Takahashi H, Nakamura K, Kaneko F, Nakagawa H, Iizuka H. Analysis of psoriasis patients registered with the Japanese Society for Psoriasis Research from 2002-2008. J Determol. 2011; 38(12): 1125-129.
- Atabati H, Esmaeili SA, Saburi E, Akhlaghi M, Raoofi A, Rezaei N, et al. Probiotics with ameliorating effects on the severity of skin inflammation in psoriasis: Evidence from experimental and clinical studies. J cell physiol. 2020; 235(12): 8925-937.
- Eppinga H, Poortinga S, Thio HB, Nijsten TE, Nuij VJ, van der Woude CJ, et al. Prevalence and phenotype of concurrent psoriasis and inflammatory bowel disease. Inflamm Bowel Dis. 2017; 23(10): 1783-789.
- Merola JF, Li T, Li WQ, Cho E, Qureshi AA. Prevalence of psoriasis phenotypes among men and women in the USA. Clinic Exper Dermatol. 2016; 41(5): 486-89.
- Guinot C, Latreille J, Perrussel M, Doss N, Dubertret L. Psoriasis: characterization of six different clinical phenotypes. Experiment Dermatol. 2009; 18(8): 712-19.
- Schön MP. Animal models of psoriasis: a critical appraisal. Experimental dermatology. 2008; 17(8): 703-12.
- Cai Y, Fleming C, Yan J. New insights of T cells in the pathogenesis of psoriasis. Cell Mol Immunol. 2012; 9(4): 302-309.
- Martins AM, Ascenso A, Ribeiro HM, Marto J. The Brain–skin connection and the pathogenesis of psoriasis: a review with a focus on the serotonergic system. Cells 2020; 9(4): 796.
- Dand N, Satveer K, Francesca C, Catherine H. S, Simpson MA, et al. Psoriasis and genetics. Acta Dermato-Venereologica 2020; 100(3): 30.
- Hugh JM, Weinberg JM. Pathophysiology of psoriasis/novel pathways, in advances in psoriasis. Springer; 2021(1): 9-18.
- Nedoszytko B, Szczerkowska-Dobosz A, Stawczyk-Macieja M, Owczarczyk-Saczonek A, Reich A, Bartosiñska J, et al. Pathogenesis of psoriasis in the “omic” era. Part II. Genetic, genomic and epigenetic changes in psoriasis. Advances in Dermatology and Allergology 2020; 37(3): 283.
- Esmaeili A, Zamani Taghizadeh Rabe S, Mahmoudi M, Rastin M. Frequencies of HLA-A, B and DRB1 alleles in a large normal population living in the city of Mashhad, Northeastern Iran. Iran J Basic Med Sci. 2017; 20(8): 940.
- Ghodke Y, Joshi K, Chopra A, Patwardhan B. HLA and disease. Euro J Epidemiol. 2005; 20(6): 475-88.
- Sharifi A, Ghadiri A, Salimi A, Ghandil P, Esmaeili S. Evaluating the distribution of (+ 2044G/A, R130Q) Rs20541 and (-1112 C/T) Rs1800925 polymorphism in IL-13 gene: An association-based study with asthma in Ahvaz, Iran. Int J Med Lab. 2021; 8(1): 62-9.
- Chen L, Tsai TF. HLA‐Cw6 and psoriasis. Br J Dermatol. 2018; 178(4): 854-62.
- Raychaudhuri SK, Maverakis E, Raychaudhuri SP. Diagnosis and classification of psoriasis. Autoimmunity Rev. 2014; 13(4-5): 490-95.
- Dogra S, Bishnoi A. Childhood psoriasis: What is new and what is news. Indian J Paediatr Dermatol. 2018; 19(4): 308.
- Fry L, Baker BS, Powles AV. Psoriasis- A possible candidate for vaccination. Autoimmun Rev. 2007; 6(5): 286-89.
- Sladek R, Rocheleau G, Rung J, Dina C, Shen L, Serre D, et al. A genome-wide association study identifies novel risk loci for type 2 diabetes. Nature 2007; 445(7130): 881-85.
- Samadi-Khouzani A, Parizi PK, Ghafari F, Esmaeili SA, Peymani M, Momtazi-Borojeni AA. Association between rs619586 (A/G) polymorphism in the gene encoding lncRNA-MALAT1 with type 2 diabetes susceptibility among the Isfahan population in Iran. Int J Diabet Dev Countr 2021; 26(1): 1-5.
- Zeng J, Luo S, Huang Y, Lu Q. Critical role of environmental factors in the pathogenesis of psoriasis. J Dermatol. 2017; 44(8): 863-72.
- Peralta C, Hamid P, Batool H, Al Achkar Z, Maximus P. Psoriasis and metabolic syndrome: comorbidities and environmental and therapeutic implications. Cureus 2019; 11(12): 6369.
- Unissa R, Kumar PM, Pasha M, Begum S, Maheswari B. Psoriasis: a comprehensive review. Asian J Res Pharmaceut Sci. 2019; 9(1): 29-33.
- Enamandram M, Kimball AB. Psoriasis epidemiology: the interplay of genes and the environment. J Investig Dermatol. 2013; 133(2): 287-89.
- Knight J, Spain SL, Capon F, Hayday A, Nestle FO, Clop A. Conditional analysis identifies three novel major histocompatibility complex loci associated with psoriasis. Human Mol Gen. 2012; 21(23): 5185-192.
- Sabat R, Philipp S, Höflich C, Kreutzer S, Wallace E, Asadullah K, Volk HD, Sterry W, Wolk K. Immunopathogenesis of psoriasis. Experiment Dermatol. 2007; 16(10): 779-98.
- Chandra A, Lahiri A, Senapati S, Basu B, Ghosh S, Mukhopadhyay I, et al. Increased risk of psoriasis due to combined effect of HLA-Cw6 and LCE3 risk alleles in Indian population. Sci Rep. 2016; 6(1): 1-8.
- Diani M, Altomare G, Reali E. T cell responses in psoriasis and psoriatic arthritis. Autoimmun Rev. 2015; 14(4): 286-92.
- Pasch MC. Nail psoriasis: a review of treatment options. Drugs 2016; 76(6): 675-705.
- Albanesi C. Immunology of psoriasis. Clinic Immunol. Elsevier; 2019: 871-78.
- Dadwal A, Mishra N, Narang RK. Novel topical nanocarriers for treatment of psoriasis: An overview. Cur Pharmaceutic Design. 2018; 24(33): 3934-950.
- Mallon E, Newson R, Bunker CB. HLA-Cw6 and the genetic predisposition to psoriasis: a meta-analysis of published serologic studies. J Invest Dermatol. 1999; 113(4): 693-65.
- Borroni RG, Costanzo A. HLA‐C* 06 and psoriasis: susceptibility, phenotype, course and response to treatment. Br J of Dermatol. 2018; 178(4): 825.
- Szczerkowska-Dobosz A, Rebala K, Szczerkowska Z, Witkowska-Tobola A. Correlation of HLA-Cw* 06 allele frequency with some clinical features of psoriasis vulgaris in the population of northern Poland. J Appl Genet. 2004; 45(4): 473-76.
Type of Study: Research |
Subject:
Genetics/ Biotechnology
Received: 2021/10/30 | Accepted: 2022/05/8 | Published: 2022/06/30
Received: 2021/10/30 | Accepted: 2022/05/8 | Published: 2022/06/30
| Rights and permissions | |
 |
This work is licensed under a Creative Commons Attribution-NonCommercial 4.0 International License. |