The incidence of recurrent pregnancy loss (RPL) is reported to be between 15-20% worldwide [1, 2]. Many factors, such as fetal chromosomal abnormalities, maternal infection, chronic diseases such as diabetes and thyroid disease, alcohol consumption, tobacco use, and drugs, are involved in RPL [3-7]. Unfortunately, only 50% of cases have a known cause [8, 9]. Uncertain causes can be infections caused by intracellular bacteria. One of the intracellular bacteria is Waddlia Chondrophila (W. Chondrophila), which belongs to Chlamydiales. This bacterium is known to be an important factor in cattle miscarriages. It has also been observed in human miscarried samples [8, 10].
The potential etiological factors cause RPL and infection in the female reproductive tract. Therefore, it is important to establish a statistical relationship between infection with this bacterium and the RPL and spontaneous miscarriage. It is also complicated to detect these intracellular bacteria due to their inability to grow in the laboratory environment. For this reason, new molecular methods are needed to detect this bacterium [11]. Recently, with the advancement of the technique, diagnostic pathways have been shifted to Real-Time polymerase chain reaction (PCR), which has greatly assisted in the detection of W. Chondrophila. The 16SrRNA gene-based Real-Time PCR was developed in 2009 by Goy et al. [12]. This method can be replaced by the time-consuming, challenging, and low-efficiency methods commonly used, and the speed and ability to count is an essential feature for the study of pathogenesis. recA gene is fully conserved and more distinct than 16SrRNA [13, 14]. Therefore, the Real-Time PCR method can help detect this bacterium. Also, no information is available on the presence of this bacterium in Yazd city and Iran. The purpose of this study was evaluation of W. Chondrophila in women with RPL using Taqman Real-Time PCR and healthy childbirth in order to determine the association between RPL and the presence of this bacterium.
Materials and Methods
Sample Collection
One hundred forty-six suspected clinical specimens from 100 women (54 were controls and 46 were women with a history of RPL, ranging from 20–47 years), including vaginal swabs from both groups (n=100), and fetal tissue from women with a history of RPL (n=46), were collected. The sample swabs are performed using sterile Dacron swabs. Fetal specimens are stored in a phosphate-buffered saline (PBS) buffer medium and the transitional medium for subsequent steps. Clinical samples of women with a history of RPL and normal childbirth were obtained from Shahid Sadoughi Hospital in Yazd city of central Iran. The study was approved by the Ethics Committee of Shahid Sadoughi University of Medical Sciences, Yazd, Iran (Code: IR.SSU.REC. 1401.041).
DNA extraction
DNA was extracted from all samples according to the manufacturer's instructions with a DNA isolation kit I (Roche). First, cells are lysed, and proteins are digested by incubation with proteinase K. A special lysis/binding buffer and magnetic glass particles (MGP) are added. Finally, the purified DNA is eluted in 50 μl of a special buffer and stored at −20 °C until laboratory analysis.
Primer design
Using Primer3 software (Rozen and Skaletsky, 2000), a forward primer WadF (5'- CGGCT ACTGTTCTGTATC -3'), and a reverse primer WadR (5'- GCGTATAACCCTTTGCTTA – 3') and a Taqman probe (5'- CGGCTACTGTTC TGTATC -3') were selected to amplify a 184 bp fragment of W. Chondrophila. The region selected was 100% conserved among Waddliaceae. Primers were also blasted against the nucleotide database of the NCBI website to ensure the absence of significant homology with other microorganism sequences.
W. Chondrophila plasmid control
In order to construct positive control, the plasmid was designed by Gene Fanavaran. The W. Chondrophila sequence (GenBank accession No., CP001928.1) was synthesized and cloned into PUC57 between EcoRI and XbaI restriction enzyme sites (Biomatik, Canada). The constructed PUC57/ W. Chondrophila plasmid was confirmed by enzyme digestion and sequencing (data not shown). This vector was transformed into the Escherichia coli DH5α strain. Finally, the plasmid was extracted by the YTA plasmid extraction kit (Yekta Tajhiz Azma, Iran) and used as a positive control. The target sequence was amplified with WadF and WadR primers. The final 25 µl reaction mixture contained 0.4 mM of each primer, 1 U/ml Taq DNA polymerase (Biorad), 5 µl of PCR buffer containing (20 mM Tris/HCl, 100 Mm KCl, 3 mM MgCl2, 400 mM dNTPs) and 1 µl W. Chondrophila DNA. PCR was performed in according to the following procedure: 5 min at 95 °C, 40 cycles at 95 °C for the 20s, 55.5 °C for 20s, 72 °C for 20s. PCR products were then purified with Gel DNA Recovery Kit (Zymo Research). Extraction of recombinant plasmid DNA was performed with Plasmid Miniprep Kit (Zymo Research), and the presence of the inserted gene was confirmed by sequencing (Data not shown). Plasmids were then linearized and quantified with a NanoDrop ND-1000 Spectrophotometer. Copy numbers of the cloned gene were derived from the molecular weights of the cloning vector and inserted, diluted in 10 mM Tris–HCl, pH 8.0 to generate standards ranging from 101 to 106 molecules and stored at 20 °C. Extracted DNA was re-suspended in 50 µL of elution buffer and stored at -20 °C. To analyze all vaginal samples (aborted and controls) by Taqman Real-Time PCR, we used 1 µL of each DNA.
Real-time PCR assay development and screening
PCR product amplification and detection were performed with a real-time PCR cycler system (Rotor-Gene, Germany) for 40 cycles. To determine the optimal concentrations of the primers and probes, various concentrations of the primer and probe sets were evaluated using a Real-Time PCR assay. The optimal concentration of primers and probe was assessed with 0.5 µM and 0.025 µM by defining the one that gave the highest recorded fluorescence and the lowest threshold cycle (CT) that can be defined as the point at which the fluorescence crosses the threshold. The optimal Real-Time PCR efficacy was obtained using a cycling profile that included an initial denaturation at 95 °C for 3 min, then 40 cycles of 10 s at 95 °C and 10 s at 60 °C.
The positive plasmid was used to determine the sensitivity limits and the reproducibility of the Real-Time PCRs. A standard curve was generated for each Real-Time PCR by serial dilutions ranging from 101 to 106 copies of plasmid/ µL. To assess for possible false-negative results (linked to PCR inhibitors), an inhibition test was systematically performed on all veterinary samples (3 µL of genomic DNA and 1 µL of each positive control at a concentration of 100 copies). In all experiments, each PCR run included a negative extraction control (sterile water) containing 5 µL Diethylpyrocarbonate (DEPC) treated H2O to detect any possible contaminating DNA.
Detection of the amplification product
Ten microliters of each PCR product were electrophoresed on 2.5% agarose gels and then observed under UV illumination.
Determination of analytical specificity, sensitivity, and reproducibility of the Taqman Real-time PCR
The specificity of the Real-Time PCR was tested using DNA extracted from different bacteria commonly found in cases of vaginitis, such as Chlamydia, Klebsiella pneumonia, Campylobacter Staphylococcus aureus, Mycoplasma, and Legionella. Using the positive control plasmid, the analytical sensitivity and the reproducibility of the Real-Time PCR were assessed on duplicates with 10- fold dilutions (101–106 copies/reaction). These dilutions were used as quantification standards to construct the standard curve by plotting the plasmid copy number against the corresponding CT values through which we know the number of copies/ml in the different test samples. The DNA extracted from all the samples was examined for the presence of Waddlia Chondrophila by the same method.
Statistical analysis
All statistical analyses were performed using SPSS software for windows (Statistical Package for the Social Sciences, version 19, SPSS Inc., Chicago, Illinois, USA). Statistical analyses of the qualitative data were performed with x2 test to identify associations. To assess the reproducibility, the mean CT of duplicates obtained in 15 independent runs was compared for each concentration of plasmid DNA. Mean, standard deviation, and coefficient of variation was calculated in Graphpad.
Results
The expected band of W. Chondrophila was observed and determined to be 184 bp (Fig. 1). The specificity of the Real-Time PCR assay was 100% when testing DNA extracted from microorganisms noted in the method, indicating that the specificity of probes guaranteed a high discrimination degree between the amplicons of the two target bacteria and those of the other tested ones.
The standard curve of W. Chondrophilais shown in Figures 2A and 2B reveals a linear trajectory over 6 logs of plasmid concentration (101 to 107 copies/µL) and curve amplification with a correlation coefficient of 0.998. The average difference between ten-fold dilutions was 3.12 cycles when testing the W. Chondrophila plasmid (Figure 2C). The efficiency of the Real-Time PCR was found to be 99% for W. Chondrophilaon Rotor-Gene system (Fig. 1 A). Further, it has been shown that Real-Time PCR could detect 101 to 107 copies of plasmid (Fig. 1 A). Fifteen of 25 replicates (60%) were positive with a W. Chondrophila plasmid positive control concentration of 1 copy/µL. We determined that the limit of detection per reaction that could be identified with a 95% probability was 9 10.2 copies/reaction for W. Chondrophila. No positive samples were detected in this work.
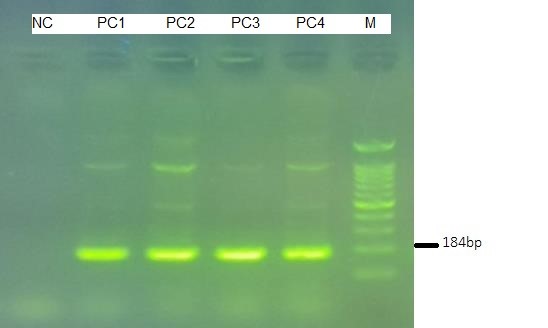
Fig. 1. Gel electrophoresis of Real-Tme PCR of W. Chondrophila positive control. NC= Negative Control; PC1-4: Positive Samples, M: Molecular weight. Considering the creation of a band within 100-200bp, which was expected based on the primer's potential product, and the repetition of this result in 4 steps (bands PC1-4) and the absence of a false band, Real-Time PCR performance has been confirmed by gel electrophoresis method.
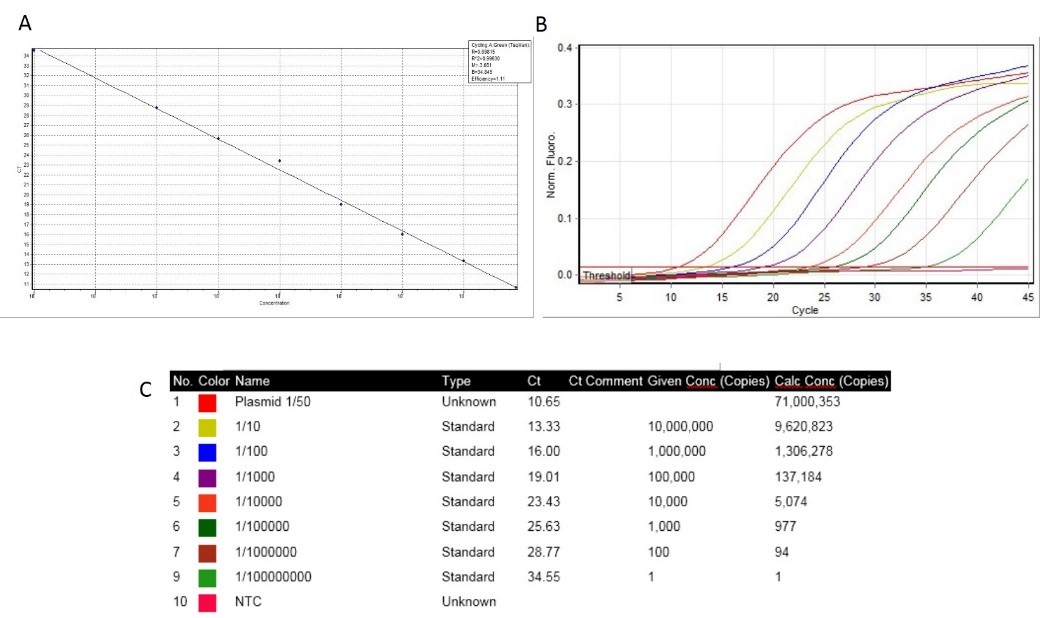
Fig. 2. The standard curve of W. Chondrophila. In this study, CT values less than 25 were considered positive, so concentrations higher than 1/10000 can be measured using the Real-Time PCR method designed in this study.
Discussion
RPL is one of the most complex and unpleasant consequences of pregnancy. The exact cause of RPL has not yet been entirely determined. Intracellular bacterial W. Chondrophila is a major cause of RPL in cattle [15] and other mammals such as humans. It has the potential for RPL in pregnant women [10]. However, its association with RPL has not yet been fully established. Infection detection methods are still evolving, and new findings are being made daily to make diagnosis more manageable and more effective in controlling the infection. Methods of amplification of nucleic acids can be performed on high-sensitivity multiple-stranded specimens that are economically viable.
Diagnosis of the bacterium W. Chondrophila is important. This bacterium has no known transmission method; it seems to be found in biological specimens such as serum, feces, etc. However, since these microorganisms are difficult to isolate from the culture medium, it is important to find new diagnostic methods because of their low concentration and resistance to the culture medium. Nowadays, replication methods such as PCR are used to detect W. Chondrophila. Finally, with the evolution of quantitative methods, the path was shifted to Real-Time PCR. This study aimed to determine the presence of W. Chondrophila in women with miscarriage and healthy delivery to determine the relationship between RPL and the presence of this bacterium using Real-Time PCR. In this study, all collected samples were negative for W. Chondrophila.
A 2007 study by Baud et al. investigated the role of W. Chondrophila in the prevalence of RPL in women with RPL, and RPL was determined using Western blotting and anti-Waddlia antibody reactivity by immunofluorescence. According to this study, there is a strong relationship between the presence of W. Chondrophila-specific IgG antibodies and early fetal loss [10]. In 2008, Siemer and colleagues investigated Chlamydia trachomatis infection as a risk factor for infertility in women in West Africa. Real-Time PCR analyzed urine samples, and the enzyme-linked immunosorbent assay (ELISA) tests were performed on their serum to determine the levels of IgG and IgA antibodies. Overall, 1.6% of infertile women were positive for Chlamydia trachomatis, but similar studies found the diagnostic value and sensitivity of the ELISA test for genital Chlamydia trachomatis infection to be very low; They concluded that serologic tests did not help detect chlamydial infections of the genital tract and trachoma [16].
In 2012, Kebbi-Beghdadi and colleagues examined the detection of W. Chondrophila by immunogenic proteins. They were looking for reliable, high-throughput serological methods. To do this, they used a combination of genomic and proteomic methods. They used two new proteins, Wim3 and Wim4 (recombinant proteins expressed in Escherichia coli), as antigens in an ELISA method. They concluded that these immunogenic proteins could be used in serological tests [17].
In 2014, Baud and his colleagues investigated the role of W. Chondrophila infection in the placenta and its association with RPL using Real-Time PCR and immunohistochemically methods. Their results showed a strong association between W. Chondrophila infection and RPL in women. They suggested that when we suspect RPL associated with W. Chondrophila, it is recommended that Real-Time PCR be performed on the placenta and vaginal swab specimens [18].
Baud et al. in Switzerland investigated the effects of sperm infection on infection with W. Chondrophila using methods such as immunohistochemistry and Real-Time PCR. In this study, sperms were artificially infected. They reported that this bacterium significantly negatively affected the function and structure of human sperm DNA [19].
Conclusion
In this study, the Real-time PCR method to measure W. Chondrophila was optimized in terms of sensitivity and specificity. This method allowed the detection of Waddlia DNA. Given the evidence that this bacterium is an important contributor to RPL, Taqman Real-Time PCR was inserted for 146 vaginal swabs and fetal tissue samples. However, W. Chondrophila DNA was not detected in either of the samples with RPL. Therefore, it can be concluded that Taqman Real-Time PCR is a highly sensitive and specific method
for detecting W. Chondrophila in clinical development. This was the first study in relationship with W. Chondrophila; therefore, we had more limitations, including the presence of standard strain for the clinical evaluation and the presence of specific tests such as serology and standard molecular kits. Therefore, it should use more specimens because of this bacterium's low frequency. Also, Taqman's Real-Time PCR method in this study was high sensitivity, but it is needed for more specimens and the use of other genes of this bacterium in the same methods.
Conflict of Interest
The authors declare no conflict of interest.
Acknowledgment
We would like to thank the friendly cooperation of Dr. Hadi Nadushan and Dr. Ahmed Hanarjo and the respected professors of Jahrom Azad University and Shahid Sadoughi.
References
- Kaandorp SP, Lauw MN, van der Schoot CE, Goddijn M, van der Veen F, Koene HR, et al. Prevalence of JAK2V617F mutation in women with unexplained recurrent miscarriage. Journal of Thrombosis and Haemostasis 2010; 8(12): 2837.
- Regan L, Rai R. Epidemiology and the medical causes of miscarriage. Best Practice & Research Clinical Obstetrics & Gynaecology 2000; 14(5): 839-54.
- Carp H. Progestogens and pregnancy loss Climacteric 2018; 21(4): 380-84.
- Goodarzi P, Falahzadeh K, Aghayan H, Payab M, Larijani B, Alavi-Moghadam S, et al. Therapeutic abortion and ectopic pregnancy: alternative sources for fetal stem cell research and therapy in Iran as an Islamic country. Cell and Tissue Banking 2019; 20(1): 11-24.
- Kleinman ME, Brennan EE, Goldberger ZD, Swor RA, Terry M, Bobrow BJ, et al. Part 5: adult basic life support and cardiopulmonary resuscitation quality: 2015 American Heart Association guidelines update for cardiopulmonary resuscitation and emergency cardiovascular care. Circulation 2015; 132(18): 414-35.
- Pillai RN, Konje JC, Richardson M, Tincello DG, Potdar N. Prediction of miscarriage in women with viable intrauterine pregnancy-a systematic review and diagnostic accuracy meta-analysis. European Journal of Obstetrics & Gynecology and Reproductive Biology 2018; 220: 122-31.
- Wahabi HA, Fayed AA, Esmaeil SA, Bahkali KH. Progestogen for treating threatened miscarriage. Cochrane Database of Systematic Reviews 2018; 8(1): 1-49.
- Baud D, Greub G. Intracellular bacteria and adverse pregnancy outcomes. Clinical Microbiology and Infection 2011; 17(9): 1312-122.
- Carrington B, Sacks G, Regan L. Recurrent miscarriage: pathophysiology and outcome. Current Opinion in Obstetrics and Gynecology 2005; 17(6): 591-97.
- Baud D, Thomas V, Arafa A, Regan L, Greub G. Waddlia Chondrophila, a potential agent of human fetal death. Emerging Infectious Diseases 2007; 13(8): 1239.
- Lamoth F, Pillonel T, Greub G. Waddlia: an emerging pathogen and a model organism to study the biology of chlamydiae. Microbes and Infection 2015; 17(11-12): 732-37.
- Goy G, Croxatto A, Posfay-Barbe K, Gervaix A, Greub G. Development of a real-time PCR for the specific detection of Waddlia Chondrophila in clinical samples. European Journal of clinical Microbiology & Infectious Diseases 2009; 28(12): 1483-486.
- Eisen JA. The RecA protein as a model molecule for molecular systematic studies of bacteria: comparison of trees of RecAs and 16S rRNAs from the same species. Journal of molecular evolution 1995; 41(6): 1105-23.
- Thompson C, Thompson F, Vandemeulebroecke K, Hoste B, Dawyndt P, Swings J. Use of recA as an alternative phylogenetic marker in the family Vibrionaceae. International Journal of Systematic and Evolutionary Microbiology 2004; 54(3): 919-24.
- Dilbeck P, Evermann J, Crawford T, Ward A, Leathers C, Holland C, et al. Isolation of a previously undescribed rickettsia from an aborted bovine fetus. Journal of clinical microbiology 1990; 28(4): 814-16.
- Siemer J, Theile O, Larbi Y, Fasching PA, Danso K, Kreienberg R, et al. Chlamydia trachomatis infection as a risk factor for infertility among women in Ghana, West Africa. The American Journal of Tropical Medicine and Hygiene 2008; 78(2): 323-27.
- Kebbi-Beghdadi C, Lienard J, Uyttebroeck F, Baud D, Riederer BM, Greub G. Identification of immunogenic proteins of Waddlia Chondrophila. PLoS One 2012; 7(1): 28605.
- Baud D, Goy G, Osterheld MC, Croxatto A, Borel N, Vial Y, et al. Role of Waddlia Chondrophila placental infection in miscarriage. Emerging Infectious Diseases 2014; 2(3): 460-65.
- Baud D, Vulliemoz N, Ammerdorffer A, Gyger J, Greub G, Castella V, et al. Waddlia Chondrophila, a Chlamydia-related bacterium, has a negative impact on human spermatozoa. Human Reproduction 2018; 33(1): 3-10.










































