Protein S (PS) is a vitamin K-dependent glycoprotein synthesized by endothelial and somatic cells. This protein is present in plasma both as a free molecule (40%) and as a bound molecule (60%) to C4b-binding protein (C4BP). The free form acts as an anticoagulant, while the bound form acts as an inhibitor of the complement system, thereby decreasing the level of PS in inflammatory conditions and promoting coagulation. PS serves as a cofactor for activated protein C (APC) to inactive activated factor (F) Va and FVIIIa. It also directly affects the prothrombinase complex by making it reversible, breaking apart the complex made up of FVa and FXa, which is important for regulating blood clotting [1-4]. PS deficiency has three types: type I is characterized by reduced levels of both free and total PS; type II is characterized by normal total PS levels but reduced free PS levels; and type III is characterized by normal free and total PS levels but dysfunctional PS [5]. Type I is the most frequent form, accounting for approximately 60-70% of cases, while type II and Type III are less common, representing about 25-30% and 5% of cases, respectively [6].
Most patients with mutations in the PS gene (PROS1) present with venous thromboembolism (VTE), including deep vein thrombosis (DVT) and pulmonary embolism (PE) as the main clinical manifestations [7, 8]. It has been reported that PROS1 mutations increase the risk of venous thrombosis, as well as cerebral and myocardial embolism [9, 10]. Venous thrombosis affects 1.90% of the population annually, with a median age of 29. Congenital PS deficiency is estimated to affect approximately one out of 500 individuals. However, due to its rarity and difficulty in diagnosing, the prevalence of PS deficiency in the general population remains unknown [11]. This study aimed to report the clinical and genetic characterization of four Iranian patients with PS deficiency.
Materials and Methods
Study population
The study included four patients with a PS deficiency (Pedigrees A, B, C1, and C2), with a mean age of 32.5 ± 25.73 years (two females and two males; age range: 1-64 years), from Sistan and Baluchestan province. Patients C1 and C2 were the mother and her proband, respectively. The study was approved by the Ethical Committee of Iran University of Medical Science, and written consent was obtained from all participants. The study was conducted according to the Declaration of Helsinki.
Laboratory and molecular tests
PS total antigen levels were measured using an automated instrument (Diagnostica Stago, Asnieres, France), with a reference range of 65%–140%. In the next step, genomic DNA was isolated from the peripheral blood of the patients using the Gene All Exgene Blood SV kit (Gene All Exgene Blood SV, China) and stored at –80 °C until analysis. Since there is a pseudogene for the PS gene, primers were designed under strict conditions to avoid pseudogene amplification. Subsequently, polymerase chain reaction (PCR) was performed to amplify the genomic DNA of all patients, and the resulting products were subjected to direct sequencing by the Sanger method, carried out by Pishgam BioTech Co. (Tehran, Iran). Mutations nomenclature was performed according to the Human Genome Organization recommendations (http://www. hgvs.org), and the PROS1 cDNA (NM_000313.4) and protein (NP_000304.2) sequences were retrieved from the National Center for Biotechnology Information reference sequence database.
In silico analysis
Each novel variation was subjected to in silico analysis with sorting intolerant from tolerant (SIFT; https://sift.bii.astar.edu.sg) and Polymorphism Phenotyping version 2 (PolyPhen2; http://genetics.bwh.harvard.edu/pph2) to predict the potential impact of missense variations on protein function. Additionally, Franklin (https://franklin.genoox.com) and Mutation Taster http://www.mutationtaster.org were also used for predicting the potential pathogenicity of genetic variations in both 5’ untranslated region (5’ UTR) mutation and missense variants.
Results
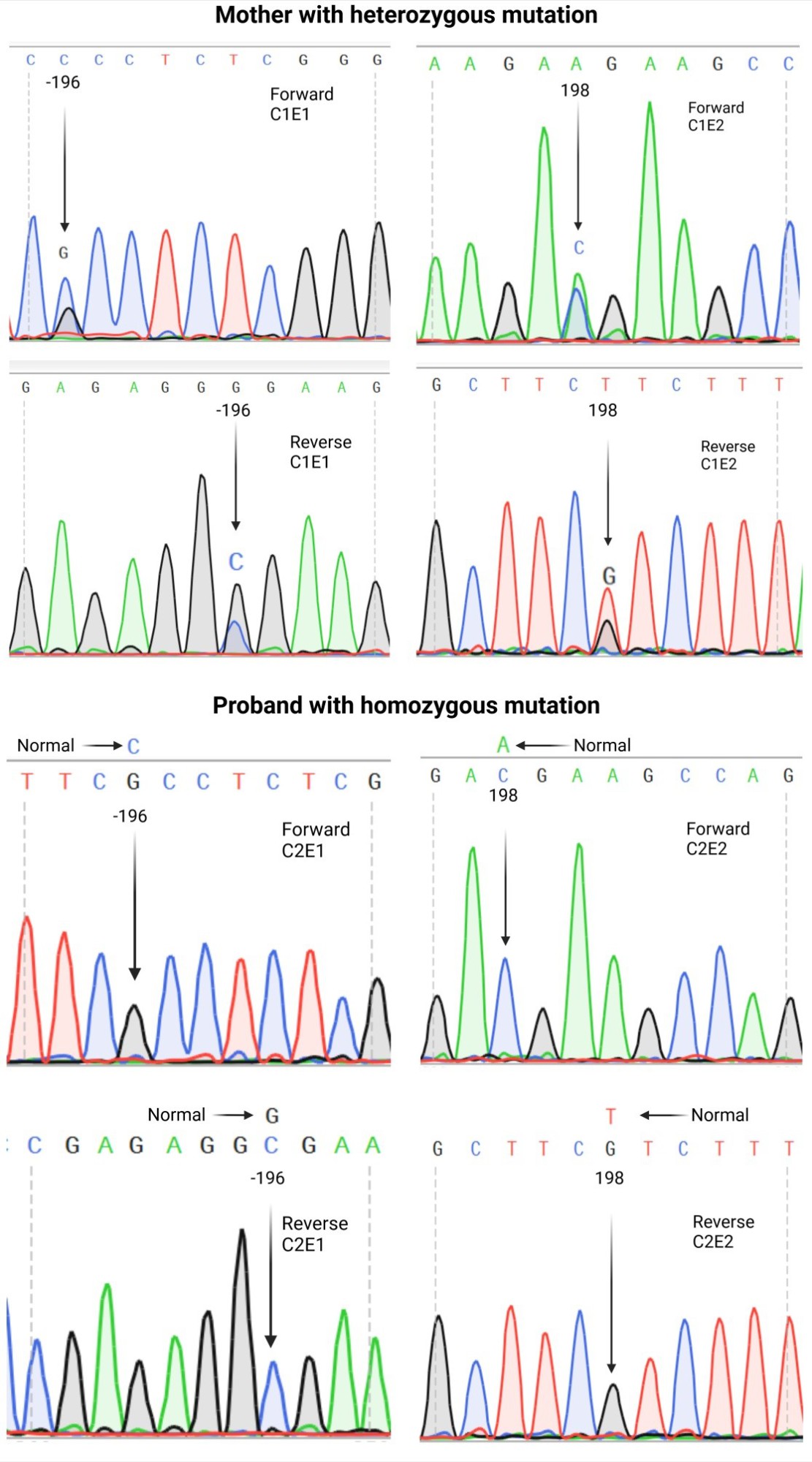
Three of the patients had a positive family history of PS deficiency and a thrombotic complication (Table 1). The mean total PS antigen level was 46.25%. In this context, patient C2 had the lowest PS level, with a total PS antigen level of 7%, and eventually experienced a fatal thrombotic complication. PROS1 variants: Variants were only detected in patients C1 and C2, who carried a missense PROS1 variant located in exon 1 (c.-196C > G) in 5’UTR and a variant in the coding region of exon 2 (c.198A > C) (Fig. 1). In this regard, these two variants were detected in a heterozygous pattern in patient C1 and a homozygous state in patient C2. To the best of our knowledge, these novel variants are being reported for the first time in two Iranian patients (Table 2).
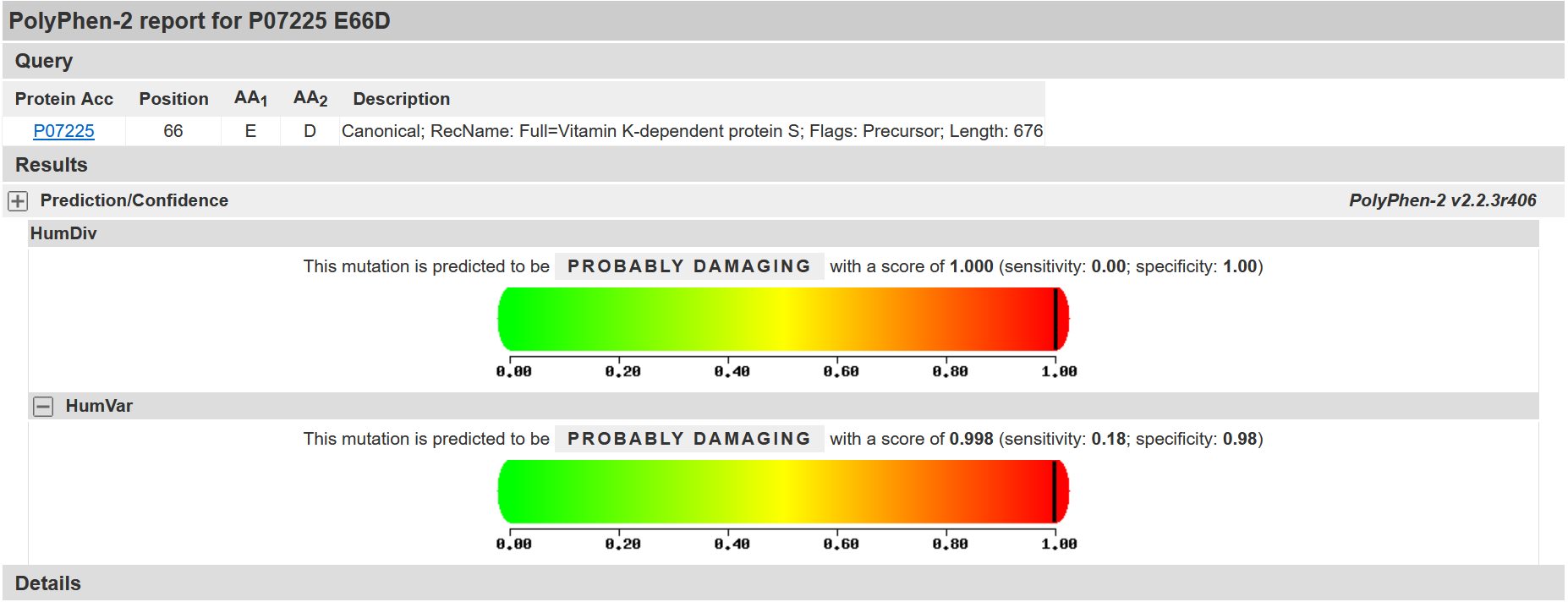
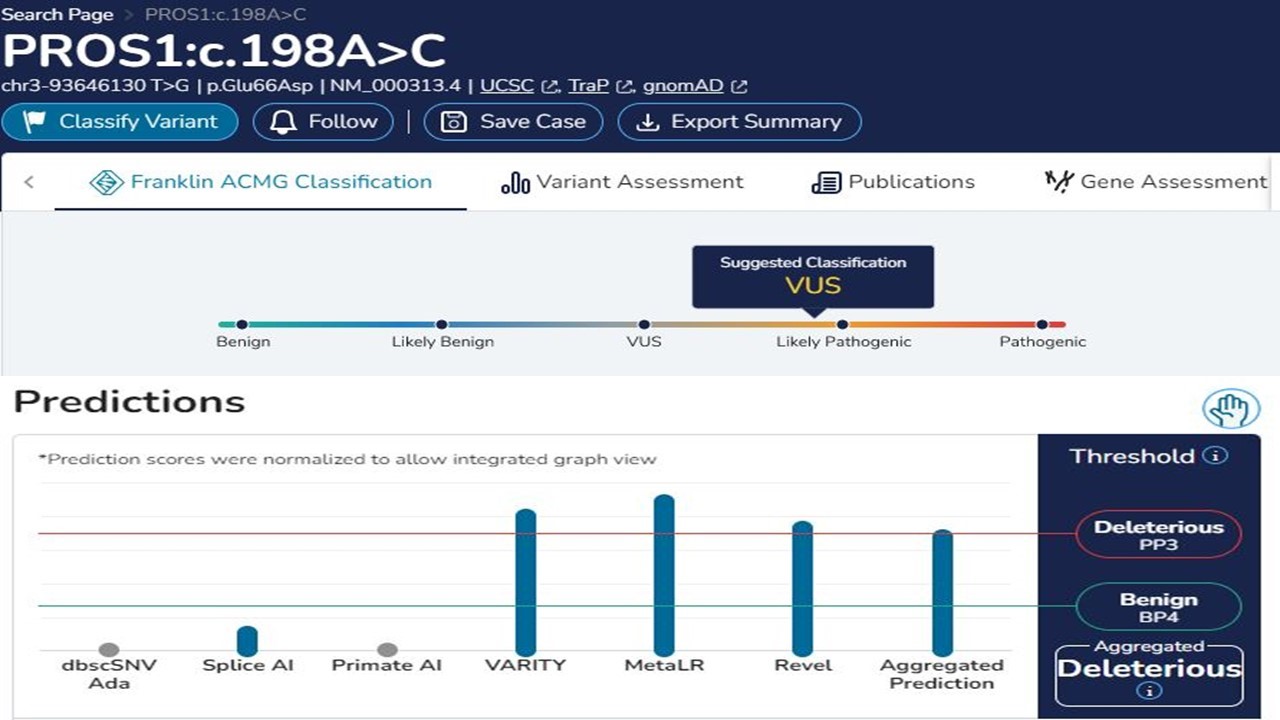
Silico analysis showed that the novel variant c.198A > C is likely detrimental. Both variants do not affect the protein’s length, but missense variant c.198A > C, which occurs in the coding region, substitutes aspartic acid with glutamic acid (p.E66D). Silico analysis of the c.198A > C variant showed that it is also a detrimental variant (PolyPhen-2 score of 1.000) (Figure 2). The result from SIFT (score: 0.00) also indicated the pathogenicity of the c.198A > C (SIFT pathogenic score: < 0.05). Furthermore, the scores computed by Franklin demonstrated the potential pathogenicity of the c.198A > C variant (Fig. 3).
Discussion
This study described four patients with PS deficiency; two of them were carrying mutations in the PROS1 gene. All patients had a lower-than-normal range of total PS antigen (65-140%), with the lowest level in patient C2. Three of the patients had a positive family history of PS deficiency and a history of thrombotic complications. It seems that co-inherence of c.-196C > G and c.198A > C variants in homozygous form in patient C2 was associated with a severe decrease in total PS antigen and subsequently early death due to thrombotic events at the age of 32 years. In contrast, the presence of these two variants in heterozygous form in patient C1 was associated with a mild decrease in PS level and a less severe clinical phenotype. Therefore, the functional effect of the c.198A>C variant is highly likely and in line with our laboratory and clinical observations.
Fig. 1. DNA sequencing (forward and reverse) of two patients with protein S deficiency. Two variants were found in patients C1 and C2. The c.-196C >G and c.198A > C variants were present in heterozygous patients in patient C1 and homozygous patients in patient C2. E1 donates exon 1, and E2 donates exon 2.
Fig. 2. In silico analysis by PolyPhen-2. Results show that the c.198A>C variant (p.Glu66Asp), located in exon 2 of the PROS1 gene, is probably to be pathogenic, as both HumDiv (1.000) and HumVar (0.998) scores fall within the pathogenic range.
Fig. 3. Franklin database suggested the c.198A>C variant in exon 2 is likely pathogenic, as the VARITY, MetaLR, Revel scores, and aggregated prediction indicate that this variant is deleterious.
Table 1. Phenotypic characteristics of Iranian patients with protein S deficiency
| Patient N. |
Age (Yrs) |
Sex |
Total protein S, % |
Consanguineous marriage of parents |
Clinical manifestation |
Management |
Family history of PSD |
Outcome |
| A |
1 |
Male |
60 |
- |
- |
Warfarin |
- |
Stable |
| B |
33 |
Female |
57 |
- |
Spontaneous bruising /Abortion/ |
Enoxaparin
Warfarin |
+ |
Stable |
| C1 |
64 |
Female |
61 |
+ |
Mild thrombotic events |
Warfarin |
+ |
Stable |
| C2 |
32 |
Male |
7 |
+ |
Severe thrombotic events |
Rivaroxaban
warfarin |
+ |
Death |
Table 2. Characteristics of novel variants found in two patients with protein S deficiency
| Patient N. |
Exon |
Nucleotide variation |
Position of protein |
Type of mutation |
Genotype |
| C1 |
1 |
c.-196C>G (chr3: 93692789G>C) |
- |
5'-UTR |
Heterozygous |
| 2 |
c.198A>C (chr3: 93646130T>G) |
p.Glu66Asp |
Missense |
Heterozygous |
| C2 |
1 |
c.-196C>G |
- |
5'-UTR |
Homozygous |
| 2 |
c.198A>C |
p.Glu66Asp |
Missense |
Homozygous |
*Patients C1 and C2 were related as mother and proband, respectively.
Our study provides further evidence for the well-established association between low levels of PS and increased risk of thrombotic events, as observed in patient C2, who suffered a fatal thrombotic event. This finding is consistent with the report by Ten Kate et al. [2], which demonstrated that lower levels of PS are associated with a higher incidence of thrombotic events. Therefore, monitoring PS levels in high-risk individuals may be a valuable strategy for identifying those at greater risk of thrombotic events.
The variant c.-196C>G was detected in the 5’ UTR region of the PROS1 gene. Silico analysis did not classify it as deleterious, but it is important to note that solely depending on this information, it is insufficient to determine the pathogenicity of a nucleotide variant. A comprehensive and detailed clinical history is necessary to accurately evaluate a mutation’s potential harm when interpreting genome-level DNA sequencing outcomes. Overall, there is no direct correlation between genotype and phenotype in patients with PS deficiency [12].
Bali et al.’s research aimed to predict the consequences of detrimental genetic mutations in the PROS1 gene and analyze its impact on DVT pathogenesis. Their findings revealed that the p.A303V variant was a crucial independent risk factor for DVT, offering valuable insights into the mechanisms of the disease. They also detected DVT in upper extremity veins, pulmonary artery, and lower extremity veins and identified a history of hypertension in eight patients. Warfarin was administered to most patients, while a few were treated with rivaroxaban [13].
In our study, patient C2 also received warfarin and rivaroxaban; however, this patient succumbed due to thrombotic events, indicating the significance of detecting PROS1 variants, as individuals with PS deficiency may require more aggressive anticoagulation to prevent thrombosis. This finding is consistent with Bali et al. [13] and highlights the significance of detecting PROS1 variants since these gene variants can result in devastating thrombotic events. In a study by Schneider et al genetic analysis of a Caucasian family suspected of hereditary PS deficiency showed a novel variant, c.1904T > C (p.Phe635Ser), which was associated with a quantitative PS deficiency. They reported that there is a possibility that the novel variant in PROS1 may cause incorrect folding and, consequently, severely impaired secretion [14].
A large-scale study by Pintao et al. [15] aimed to investigate the association between PS levels and the risk of VTE in a population-based case-control study involving 5,317 individuals. The results showed that deficiency in free and total PS levels was not significantly associated with venous thrombosis, except for a lower cutoff value of <33 U/dL of free PS levels, which showed a 5.4-fold increased risk of venous thrombosis. Importantly, they demonstrated that inherited PS deficiency was rare in the general population, as variants in the PROS1 gene were detected in only 5 out of 48 patients with very low levels of PS [15]. Accordingly, in our study, only patients C1 and C2 had mutations in PROS1 exons. Patients A and B also had PS antigen levels (60%, 57%) lower than the normal range (65-140%), and positive family history was observed in patient B. However, we could not identify these two patients’ nucleotide variations in the PROS1 gene. It should bear in mind that there could be several reasons why mutations were not found in patients A and B, such as mutations outside of exon regions or acquired deficiency due to liver disease, nephrotic syndrome, vitamin K deficiency, severe and chronic inflammation, or autoimmune syndromes. Moreover, genomic changes can be missed by direct sequencing using the Sanger method by around 10%, which could also explain the lack of mutations in patients A and B [16, 17].
It is important to note that the normal range for PS may vary among different regions and different groups of patients with varying ages and conditions. As a result, it is possible that some patients may not be diagnosed with PS deficiency based solely on the reference ranges established for a particular region. Particularly, a study conducted by Tabibian et al. investigated the influence of sex, age, oral contraceptive use, and menopause on PS plasma levels was investigated in 1200 healthy individuals aged 18-69 in Iran. Notably, the study results revealed that increasing age in females correlated with a rise in PS plasma levels. However, based on the reference ranges (Male: 91–144 IU/dL; Female: 78–120 IU/dL) from this study [18], all of our patients had PS deficiency. In this context, each region and country needs to establish its own PS reference range to manage the PS deficiency with the most accurate approach.
Conclusion
Based on the reference ranges of normal protein S activities (Male: 91–144 IU/dL; Female: 78–120 IU/dL) from this study, all the patients had PS deficiency. The results highlight the heterogeneity of PS deficiency and the need for further investigation to identify additional mutations and understand the genetic basis of this condition in the Iranian population.
Ethical Consideration
The study was approved by the Iran University of Medical Science Ethics Committee (no 1399,652), and written consent was obtained from all participants.
Funding
This study was supported by grant No. 17798 from Iran University of Medical Sciences.
Conflict of Interest
The authors approve that there is not any conflict of interest.
Acknowledgments
The authors would like to acknowledge the supportive assistance of Iran University of Medical Sciences (IUMS).
Authors’ Contributions
N.S: Laboratory works, M.S: Analysis of data. AD: Applying patient samples, Sh.T: Applying patient samples, F.Z: Project consultant, and MR. R: Manuscript editor and project manager.
References
[1]. Duebgen S, Kauke T, Marschall C, Giebl A, Lison S, Hart C, et al. Genotype and laboratory and clinical phenotypes of protein s deficiency. American Journal of Clinical Pathology 2012; 137(2): 178-84.
[2] . Ten Kate M, Van Der Meer J. Protein S deficiency: a clinical perspective. Haemophilia 2008; 14(6): 1222-228.
[3]. De Frutos PG, Fuentes-Prior P, Hurtado B, Sala N. Molecular basis of protein S deficiency. Thrombosis and Haemostasis 2007; 98(09): 543-56.
[4]. Takhviji V, Zibara K, Maleki A. A case-control study on factor V Leiden: an independent, gender-dependent risk factor for venous thromboembolism. Thrombosis J. 2021; 19(1): 74.
[5]. T Tang L, Jian XR, Hamasaki N, Guo T, Wang HF, Lu X, et al. Molecular basis of protein S deficiency in China. American Journal of Hematology 2013; 88(10): 899-905.
[6]. Beauchamp NJ, Dykes AC, Parikh N, Campbell Tait R, Daly ME. The prevalence of, and molecular defects underlying, inherited protein S deficiency in the general population. British Journal of Haematology 2004; 125(5): 647-54.
[7] . Lipe B, Ornstein DL. Deficiencies of natural anticoagulants, protein C, protein S, and antithrombin. Circulation 2011; 124(14): 365-68.
[8] . Wu Y, Liu J, Zeng W, Hu B, Hu Y, Tang LV. Protein S deficiency and the risk of venous thromboembolism in the han chinese population. Front Cardiovasc Med. 2021; 8(12): 796755.
[9]. Chan NC, Cheng CK, Chan KC, Wong CM, Lau KM, Kwong JH, et al. Distinctive regional-specific PROS1 mutation spectrum in Southern China. Journal of Thrombosis and Thrombolysis 2018; 46(1): 120-24.
[10]. Wypasek E, Potaczek DP, Płonka J, Alhenc-Gelas M, Undas A. Protein S deficiency and Heerlen polymorphism in a Polish patient with acute myocardial infarction and previous venous thromboembolism. Thrombosis Research 2013; 132(6): 776-77.
[11] . C. Dykes A, Walker ID, McMahon AD, Islam SI, Tait RC. A study of protein S antigen levels in 3788 healthy volunteers: influence of age, sex and hormone use, and estimate for prevalence of deficiency state. British Journal of Haematology 2001; 113(3): 636-41.
[12] . Richards S, Aziz N, Bale S, Bick D, Das S, Gastier-Foster J, et al. Standards and guidelines for the interpretation of sequence variants: A joint consensus recommendation of the American College of Medical Genetics and Genomics and the Association for Molecular Pathology. Genetics in Medicine 2015; 17(5): 405-23.
[13] . Bali DFA, Eroglu T, Ozkan DT. Pathogenic Ala303Val Mutation in the PROS1 Gene is Associated with the Pathogenesis of Deep Vein Thrombosis. Erciyes Medical Journal 2022; 44(2): 183-94.
[14]. Schneider S, Reißig J, Weisbach V. Protein S Erlangen: A novel PROS1 gene mutation associated with quantitative protein S deficiency. Blood Coagulation & Fibrinolysis 2022; 33(4): 224-27.
[15] . Pintao MC, Ribeiro DD, Bezemer ID. Protein S levels and the risk of venous thrombosis: results from the MEGA case-control study. Blood 2013; 122(18): 3210-219.
[16] . Mulder R, Tichelaar VY, Lijfering WM. Decreased free protein S levels and venous thrombosis in the acute setting, a case-control study. Thrombosis Research 2011; 128(5): 501-502.
[17] . Stahl CP, Wideman CS, Spira TJ, Haff EC, Hixon GJ, Evatt BL. Protein S deficiency in men with long-term human immunodeficiency virus infection. Blood 1993; 81(7):1801-07
[18] . Tabibian S, Khoshmirsafa M, Paridar M. Reference interval of antithrombin, protein C, and protein S activities in healthy adults in Iran, the effect of age, sex, oral contraceptive intake, and menopause. International Journal of Laboratory Hematology 2022; 44(3): 626-34.










































 , Mohammad Reza Rezvany *
, Mohammad Reza Rezvany * 

 , Mahmood Shams
, Mahmood Shams 
 , Akbar Dorgalaleh
, Akbar Dorgalaleh 

 , Shadi Tabibian
, Shadi Tabibian 

 , Farhad Zaker
, Farhad Zaker