The prevalence of human malignancies has risen at an alarming rate globally in the last decades [1]. Bladder urothelial carcinoma (BLCA) is one of the top ten prevalent diagnosed malignancies all over the world, with an incidence of 573,000 new cases and 213,000 deaths reported in 2020 [2]. Considerable evidence has demonstrated that cigarette smoking is the most significant risk factor for BLCA, and almost 50% of all BLCA cases are cigarette smokers [3]. Overall, more than 70% of all newly diagnosed BLCA cases are assorted as non-muscle invasive bladder cancers (NMIBCs). Most often, even if the tumors are completely removed after standard therapy, roughly 50% of NMIBCs recur, and around 10% to 15% of them progress to muscle-invasive bladder cancer (MIBC) [4]. The high recurrence rate and progression of NMIBC to MIBC remain significant challenges in urologic oncology, posing a heavy load on the healthcare systems [5]. Thus, the appraisement of novel potential targets regulating BLCA initiation and progression is required.
The isoleucine glutamine motif-containing GTPase-activating protein (IQGAP) family contains three related members in humans, IQGAP1, 2, and 3, sharing a high degree of homology and similar domain structures. These scaffold proteins have been shown to mainly regulate cellular processes such as cell division, cell adhesion, and cytoskeletal dynamics [6].
Growing attention has been paid to the role of IQGAPs in the occurrence and progression of cancers in recent years. Experimental evidence and clinical studies suggest that dysregulated IQGAPs play an essential role in tumor occurrence and advancement, and alterations in their expression are closely related to patient prognosis. IQGAP1 and IQGAP2 exert a tumor suppressor role in BLCA, whereas IQGAP3 seems to act as an oncogene that can participate in cancer growth and metastases [7]. As the newest member of the IQGAP family, IQGAP3 was discovered in 2007, and the current knowledge of IQGAP3 biology needs to catch up compared to IQGAP1 and IQGAP2 [8]. Nonetheless, accumulative evidence supports IQGAP3's oncogenic role. For instance, IQGAP3 overexpression was reported to facilitate the proliferation of bladder cancer cells, and its silencing could impressively suppress cell proliferation [9]. More importantly, a previous study showed that IQGAP3 expression was markedly elevated in tissue and urine samples of BLCA patients. Higher IQGAP3 mRNA expression was associated with higher tumor grade and worse prognosis. Therefore, IQGAP3 can be suggested as a diagnostic marker and therapeutic target for BLCA [10]. However, the oncogenic role of IQGAP3 in BLCA has yet to be fully elucidated. The current study intended to inspire a comprehensive analysis of IQGAP3 to specify its potential value in BLCA as a possible diagnostic and prognostic biomarker based on a range of large public databases (Fig. 1).
Fig. 1. The study workflow. Several bioinformatics platforms were employed to investigate the expression, promotor methylation, genetic alteration, and functional and survival analyses of IQGAP3 in bladder urothelial carcinoma.
IQGAP3= Isoleucine–glutamine motif-containing GTPase-activating protein 3.
Materials and Methods
Analysis of IQGAP3 expression
The transcription level of IQGAP3 between tumor and normal specimens across different human tumor types was comprehensively figured out using the TNMplot database [11] (https: //www.tnmplot.com accessed on 17 December 2023). The significant differences were compared via the Mann-Whitney U test and marked with red*.
Subsequently, the mRNA expression pattern of IQGAP3 was studied between normal bladder cases and BLCA cases using TNMplot (accessed on 17 December 2023) and UALCAN [12] (http: //ualcan.path.uab.edu/ accessed on 17 December 2023). Statistical analyses were computed using the U Mann-Witney and the student's t-test in TNMplot and UALCAN, respectively.
The mRNA expression pattern of IQGAP3 based on clinicopathological characteristics in BLCA was further assessed using the TCGA dataset in the UALCAN and UCSC Xena (http: //xena.ucsc.edu/ accessed on 20 December 2023) [13] web-based tools.
Promotor methylation analysis of IQGAP3 in BLCA
IQGAP3 promoter methylation level was evaluated using UALCAN and normalized as β values. Student’s t-test was employed, and p < 0.05 was considered significant.
Survival analysis of IQGAP3 in BLCA
The Kaplan–Meier Plotter [14] (www.kmplot.com/ accessed on 18 December 2023) was applied to check out the prognostic value of IQGAP3 expression in BLCA patients. The overall survival (OS) and recurrence-free survival (RFS) were represented using the Kaplan–Meier Plotter. Log-rank p-value, 95% confidence interval, and hazard ratios (HR) were determined.
Functional analysis of IQGAP3 in BLCA
The protein-protein interaction (PPI) networks of IQGAP3 and its 20 top frequently altered neighboring genes were constructed using GeneMANIA [15] (http: //www.genemania.org accessed on 17 December 2023).
The top 50 most frequently altered genes with IQGAP3 in BLCA were attained from cBioPortal (http: //www.cbioportal.org/ [16] accessed on 17 December 2023). The P-value of < 0.05 was considered as the cut-off. Further, the gene ontology (GO) (GO terms such as functional annotation, biological process, and molecular function) and Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway analysis of the top 50 genes associated with IQGAP3 were generated using Enrichr [17] (https: //maayanlab.cloud/Enrichr/ (accessed on 17 December 2023)) platform. P-value < 0.05 was set as a criterion.
Analysis of IQGAP3 genetic alteration in BLCA
The cBioPortal online tool (accessed on 17 December 2023) was utilized to inspect the genetic alteration of IQGAP3 in BLCA based on the Bladder Urothelial Carcinoma (TCGA, PanCancer Atlas) dataset, containing data from 411 patients. Moreover, the association between the commonly mutated genes and the expression of IQGAP3 was studied by MuTarget [18] (https: //www.mutarget.com/result (accessed on 19 December 2023)) platform. The condition of selected genes was FC > 1.44 and p-value < 0.01.
Correlations between IQGAP3 expression and immune genes
The correlation of IQGAP3 with immune markers in BLCA was scrutinized via TIMER2.0 [19] (http: //timer.comp-genomics.org/ (accessed on 17 December 2023)) tool using Spearman’s correlation analysis.
Results
Transcription levels of IQGAP3 in BLCA
The expression pattern of IQGAP3 in
pan-cancer was first scoured using the TNMplot database. Results illustrated that the IQGAP3 gene was appreciably overexpressed in various human tumors, including BLCA, compared to normal cases (Fig. 2A).
Then, the transcription level of IQGAP3 between BLCA and normal tissues was also appraised using TNMplot (Figure 2B) and UALCAN (Fig. 2C) databases. The findings from TNMplot (p = 1.11e-16) and UALCAN (p = 5.13e-07) elucidated that IQGAP3 transcription level was impressively elevated in BLCA versus normal specimens.
Relationship between IQGAP3 expression and clinicopathological characteristics in BLCA
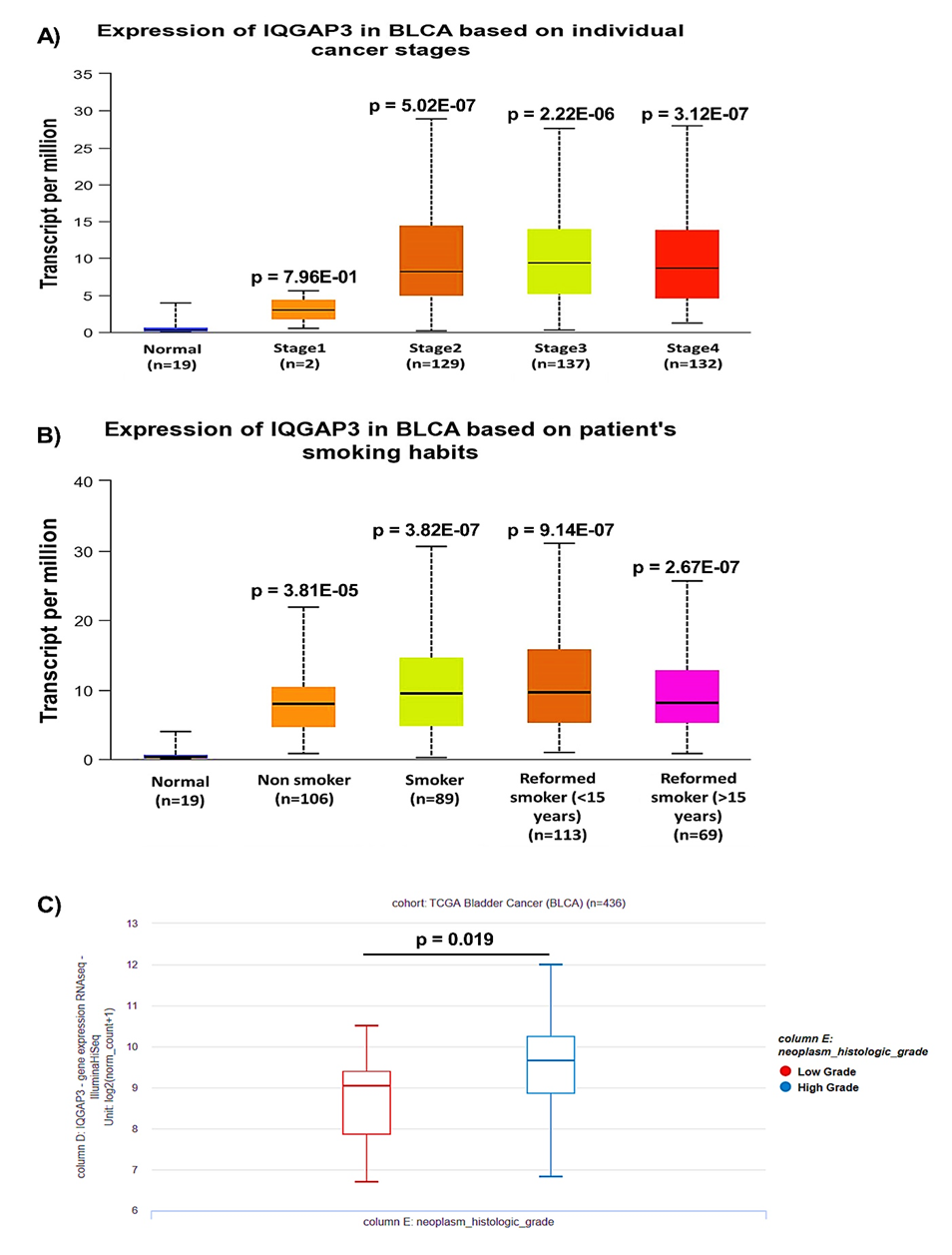
The relationship between IQGAP3 expression and clinicopathological characteristics was determined using UALCAN and UCSC Xena web-based platforms. The results from UALCAN indicated that the expression of IQGAP3 was related to advanced stages (Fig. 3A). We further executed a subgroup analysis based on smoking status. However, no meaningful change was seen between smoker patients and non-smoker patients (p = 3.65e-01) (Fig. 3B). An analysis from UCSC Xena using Welch's t-test cleared that IQGAP3 expression was remarkably elevated in high-grade tumors versus low-grade tumors (Fig. 3C; p = 0.019).
Analysis of IQGAP3 promotor methylation in BLCA
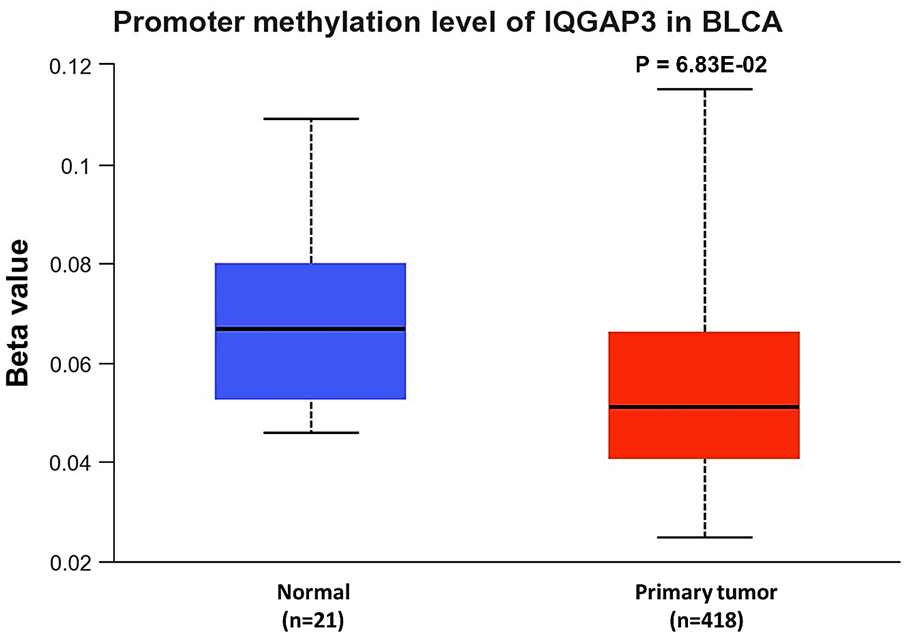
The UALCAN tool was applied to evaluate the DNA methylation status of the IQGAP3 promotor. No meaningful change was detected in IQGAP3 promotor methylation level between BLCA patients and normal individuals (Fig. 4, p = 6.83e-02).
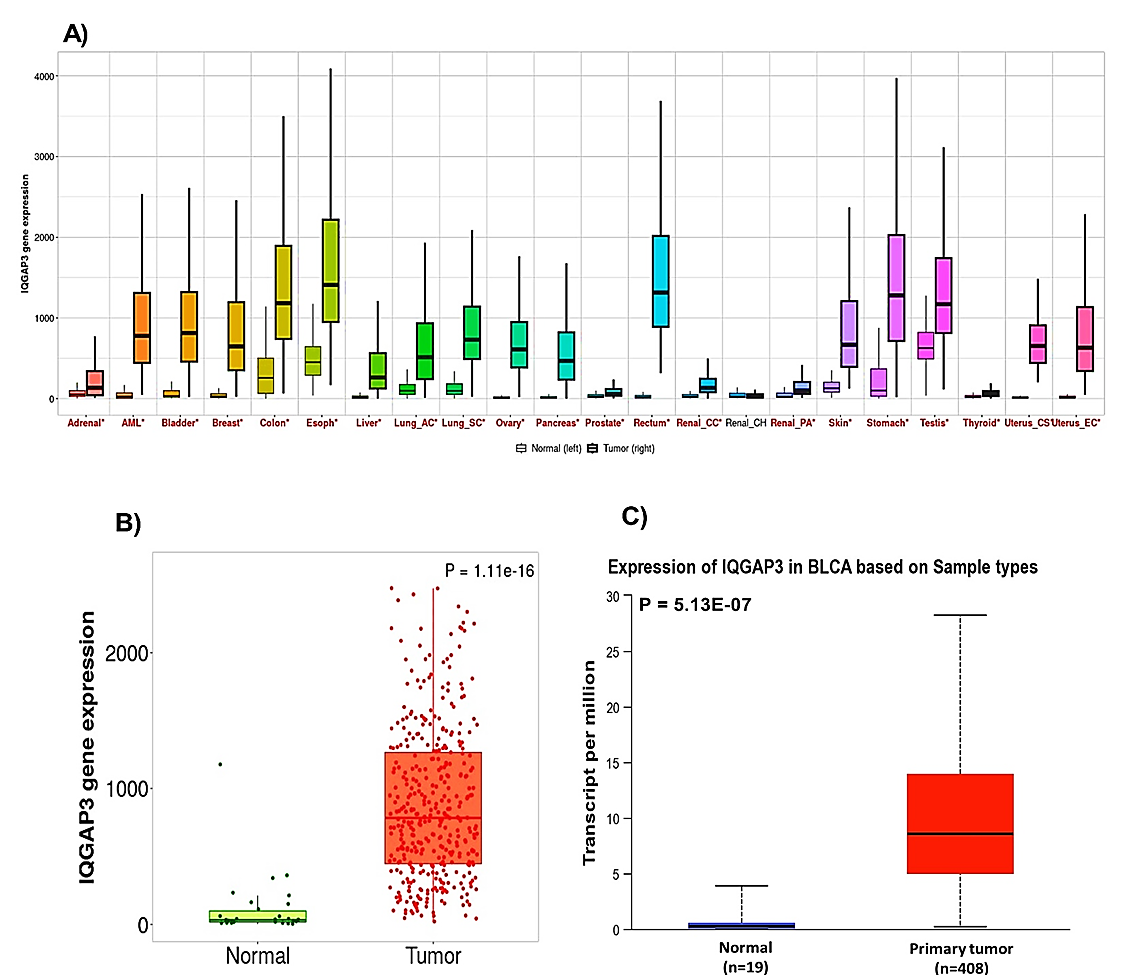
Fig. 3. Association between IQGAP3 expression and clinicopathological characteristics in BLCA patients. The relationship between IQGAP3 expression with A) tumor stage, B) smoking status, and C) tumor grade was assessed using UALCAN and UCSC Xene tools.
IQGAP3= Isoleucine–glutamine motif-containing GTPase-activating protein 3; BLCA= Bladder urothelial carcinoma; UALCAN= University of Alabama at Birmingham Cancer; UCSC= University of California, Santa Cruz.
Fig. 4. Promoter methylation analysis of IQGAP3 in BLCA using the UALCAN database. IQGAP3= isoleucine–glutamine motif-containing GTPase-activating protein 3; BLCA= Bladder urothelial carcinoma; UALCAN= University of Alabama at Birmingham cancer.
Genetic alterations of IQGAP3 in BLCA patients
The cBioPortal online platform was employed to elucidate the genetic alteration profiling of IQGAP3 in patients with BLCA using the TCGA dataset. As displayed in Figure 5A, the IQGAP3 gene was altered in 25 (6%) of 411 bladder cancer patients. Among genetic alterations in the IQGAP3 gene, amplification was the predominant alteration type (3.41%), followed by mutation (1.95%) and deep deletion (0.24%). Besides, multiple alterations were also found in 0.49% of cases (Fig. 5B).
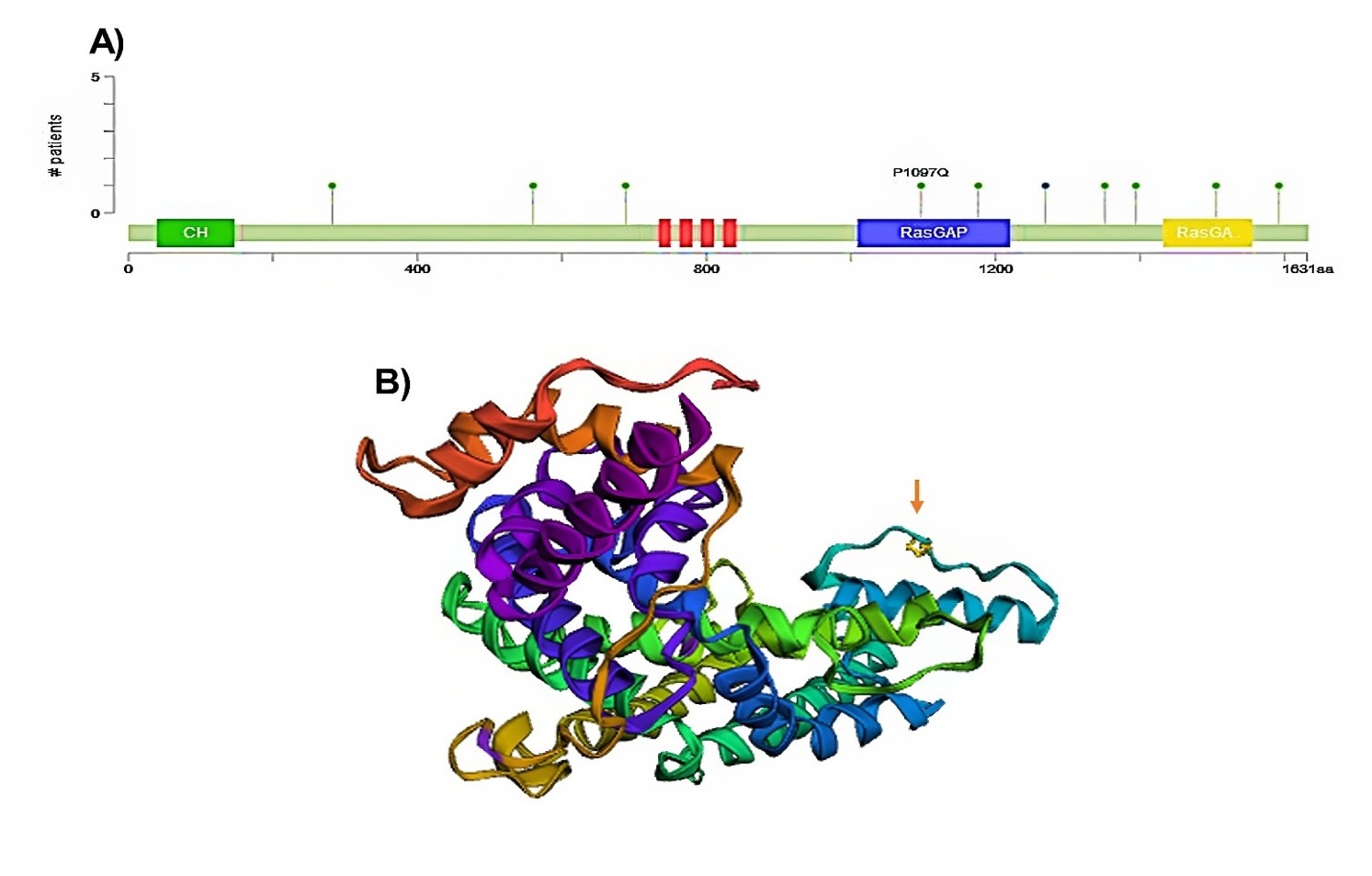
As depicted in Figure 5C, wild-type IQGAP3 had a lower expression level than the mutant type in BLCA. However, there was no statistical difference between the mutant and non-mutant types (Figure D). Further, the detailed mutation sites of IQGAP3 are represented in Figures 6A and B. In IQGAP3, ten mutation sites (including nine missense mutations and one truncation mutation) were detected, located between amino acids 0-1631, of which the amino acid number 1097 had the highest mutation frequency (Fig. 6A).
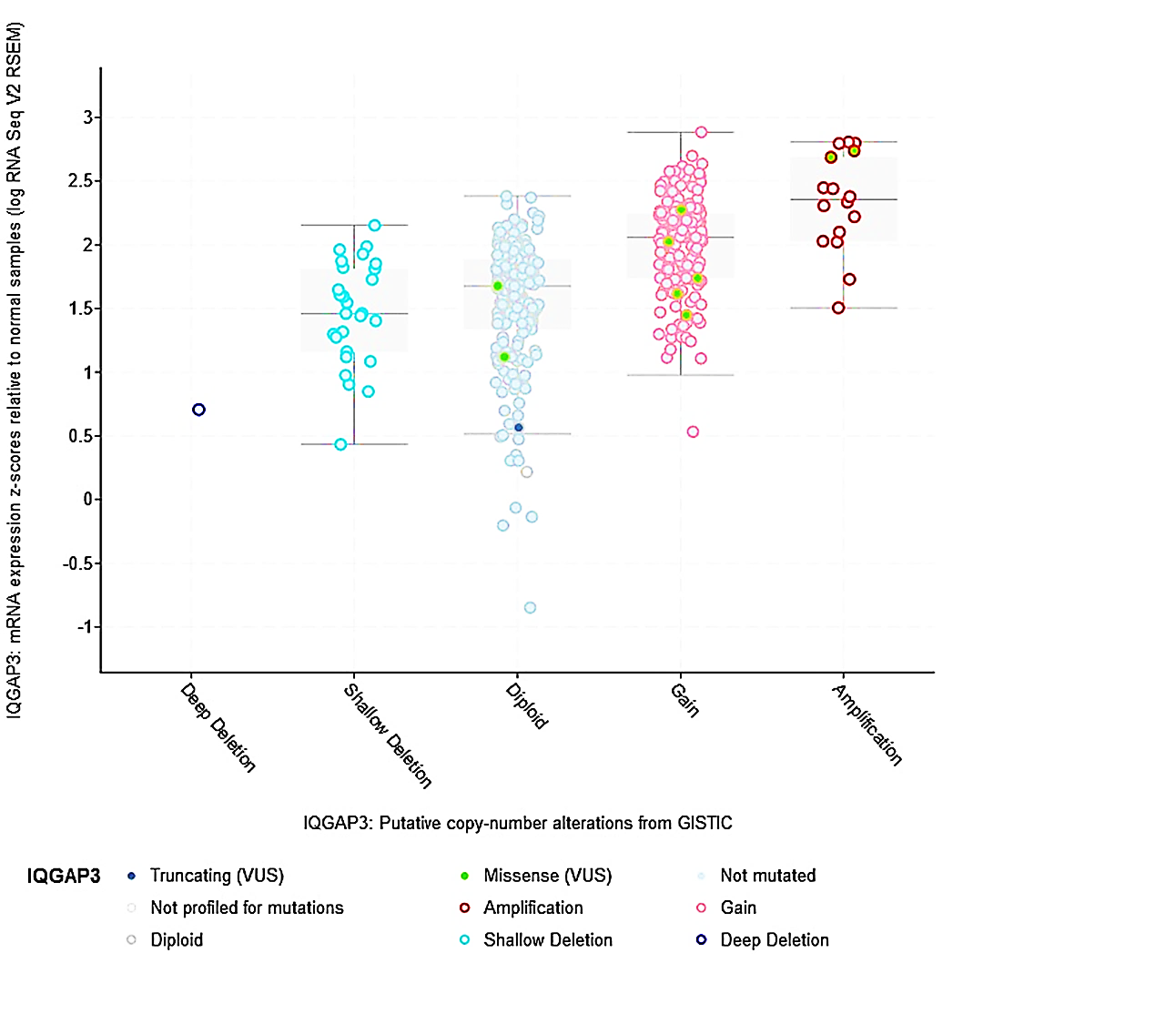
This site in the 3D structure of IQGAP3 protein is presented in Figure 6B. Then, we assessed the relation between IQGAP3 copy number and its expression in BLCA. As shown in Figure 7, the amplified type had higher expression than other types in BLCA. Finally, the MuTarget platform was used to identify crucial mutant genes responsible for IQGAP3 overexpression; as shown in Table 1, the top ten mutant genes positively correlated with the expression of IQGAP3 were RB1, TP53, ASTN2, NCKAP5, NUP205, HECTD4, ANLN, CDH9, BSN, and FLG2 in BLCA. As shown in Figure 8, the four most robustly associated mutant genes with IQGAP3 expression levels were plotted. Correlation between IQGAP3 with immune checkpoints in BLCA
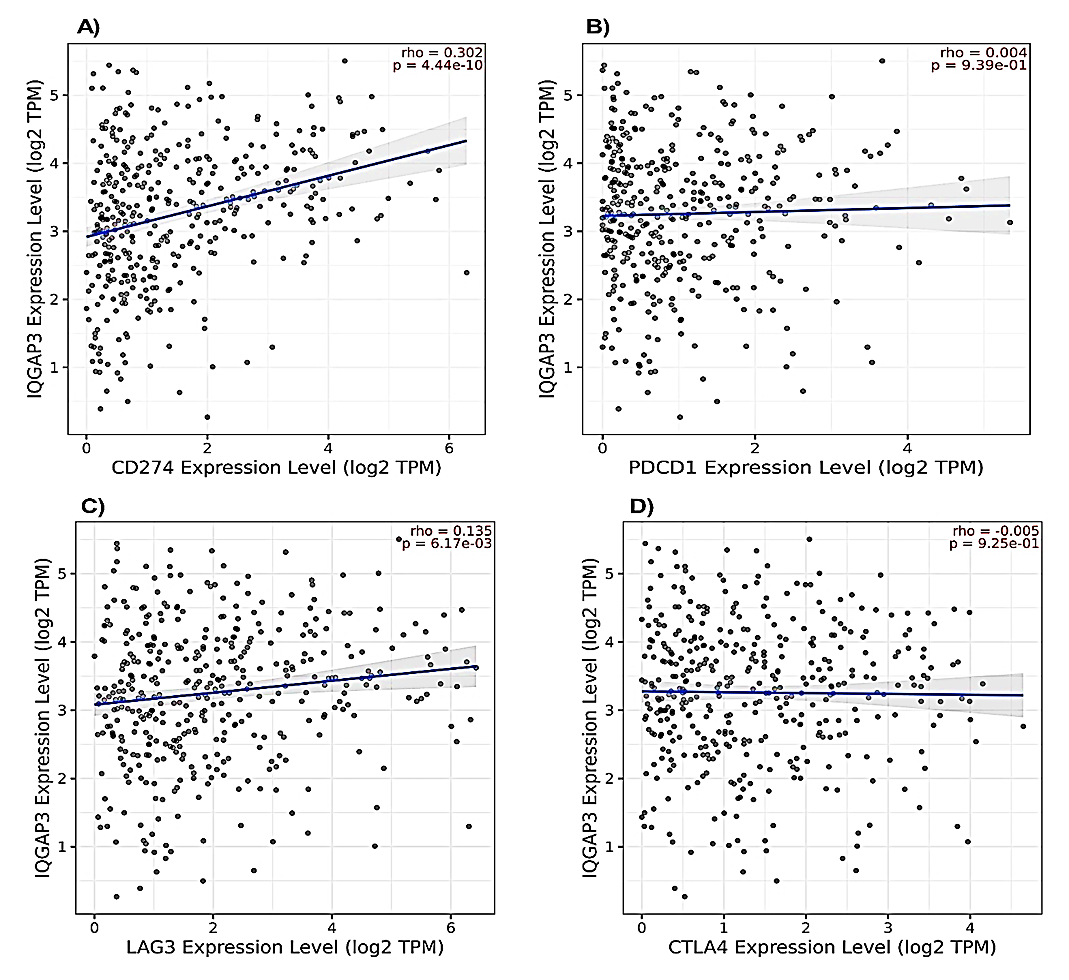
Due to the increasingly important role of immunotherapy in the treatment of bladder cancer, the relationship between the expression of IQGAP3 with PD-L1, CTLA4, LAG3, and PD-1 was further assessed using the TIMER2 platform. We found a positive correlation between IQGAP3 expression with PD-L1 (rho = 0.302, p = 4.44e−10) and LAG3 (rho = 0.135, p = 6.17e−03), while no correlation was detected between IQGAP3 expression with PD-1 (rho = 0.004, p = 9.39e−01) and CTLA4 (rho = -0.005, p = 9.25e−01) (Fig. 9A-D).
Fig. 5. Analysis of IQGAP3 genetic alterations in BLCA. A) Genetic alteration map, B) Genetic alterations frequency, C) Relationship between IQGAP3 mutation and its expression level in cBioPortal-based BLCA.
IQGAP3= Isoleucine–glutamine motif-containing GTPase-activating protein 3. BLCA= Bladder urothelial carcinoma
Fig. 6. Analysis of IQGAP3 mutation alterations in BLCA in eBioPortal database. Mutation sites on A) amino acid sequence and B) crystal structure of IQGAP3.
IQGAP3= isoleucine–glutamine motif-containing GTPase-activating protein 3. BLCA= Bladder cancer.
Fig.7. Copy number alterations of IQGAP3 in BLCA using cBioPortal.
IQGAP3= Isoleucine–glutamine motif-containing GTPase-activating protein 3; BLCA= Bladder urothelial carcinoma
Table 1. The top ten mutant genes positively related to isoleucine–glutamine motif-containing GTPase-activating protein 3 in bladder urothelial carcinoma from MuTarget
| Mutation of |
Mean expression (mutant) |
Mean expression (wild) |
Number of mutants |
Number of wild |
FC (mutant/wild) |
Direction |
P-value |
| RB1 |
1840.22 |
1115.71 |
74 |
334 |
1.65 |
up |
7.63e-11 |
| TP53 |
1492.94 |
1028.61 |
192 |
216 |
1.45 |
up |
1.01e-09 |
| ASTN2 |
2324.22 |
1222.82 |
9 |
399 |
1.9 |
up |
3.67e-04 |
| NCKAP5 |
1872.3 |
1209.77 |
23 |
385 |
1.55 |
up |
6.30e-04 |
| NUP205 |
1795.17 |
1212.86 |
24 |
384 |
1.48 |
up |
8.03e-04 |
| HECTD4 |
1837.77 |
1206.92 |
26 |
382 |
1.52 |
up |
1.12e-03 |
| ANLN |
1929.82 |
1228.2 |
11 |
397 |
1.57 |
up |
1.16e-03 |
| CDH9 |
2307 |
1223.21 |
9 |
399 |
1.89 |
up |
1.16e-03 |
| BSN |
1889.76 |
1205.17 |
25 |
383 |
1.57 |
up |
1.26e-03 |
| FLG2 |
1924.06 |
1191.45 |
31 |
377 |
1.61 |
up |
1.55e-03 |
Interaction network analysis of IQGAP3 in bladder cancer
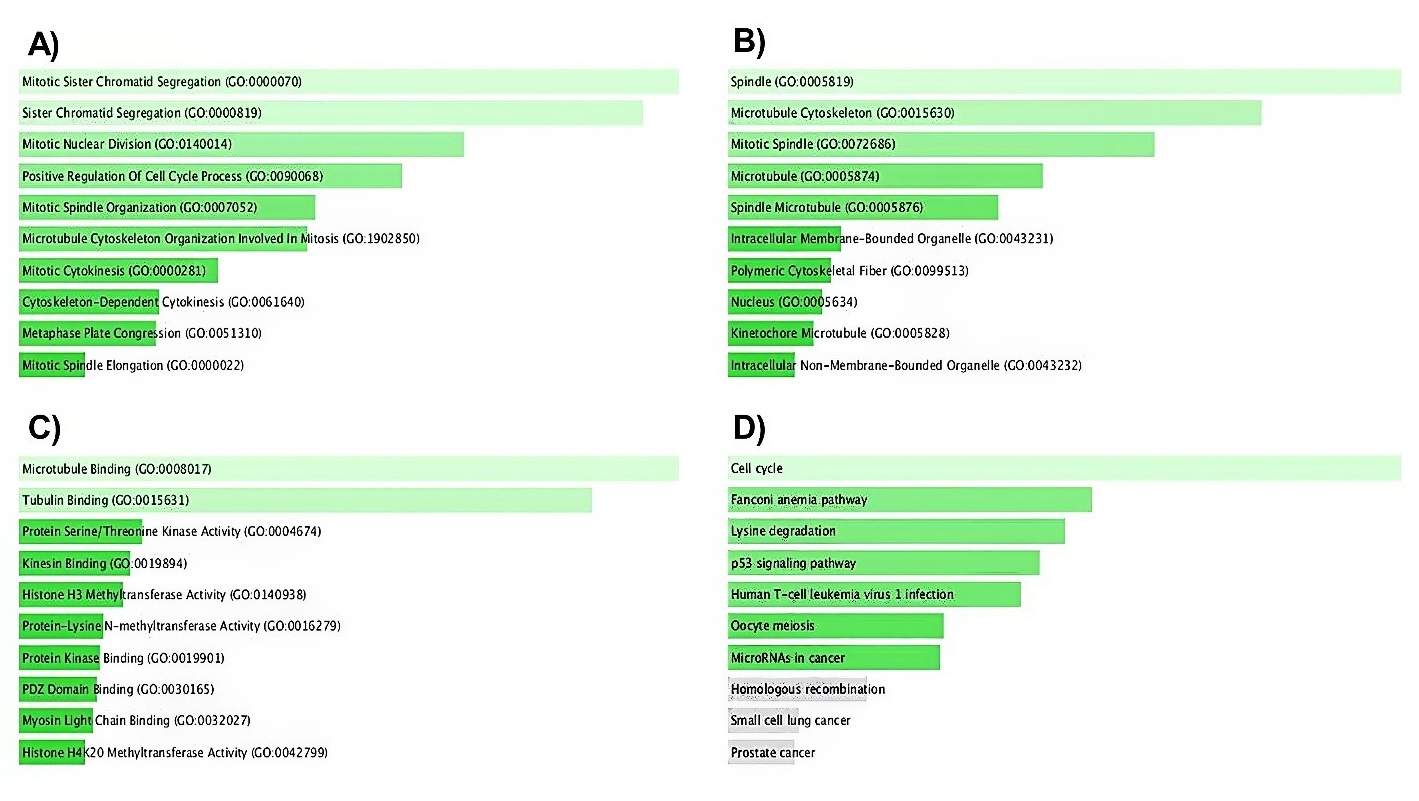
To better understand IQGAP3 molecular mechanisms in tumorigenesis, we conducted various pathway enrichment analyses on IQGAP3-interacting proteins and genes. The network of IQGAP3 and its 20 related neighboring proteins was constructed via the GeneMANIA database (Fig. 10A). The relationship between IQGAP3 expression and its most correlated gene (CDC42) is displayed in Figure 10B. Subsequently, the Enrichr database was applied to understand the GO features and signaling pathways related to IQGAP3. The most commonly enriched biological processes for IQGAP3 and its neighbor genes were mitotic sister chromatid segregation and sister chromatid segregation, respectively (Fig. 11A). The analysis of cellular components (Fig. 11B) and molecular functions (Fig. 11C) revealed that IQGAP3 and its correlated genes were primarily enriched in spindle and microtubule binding, respectively. In KEGG pathway analysis, we announced that these genes were most commonly enriched in the cell cycle (Fig. 11D).
The prognostic role of IQGAP3 in BLCA patients
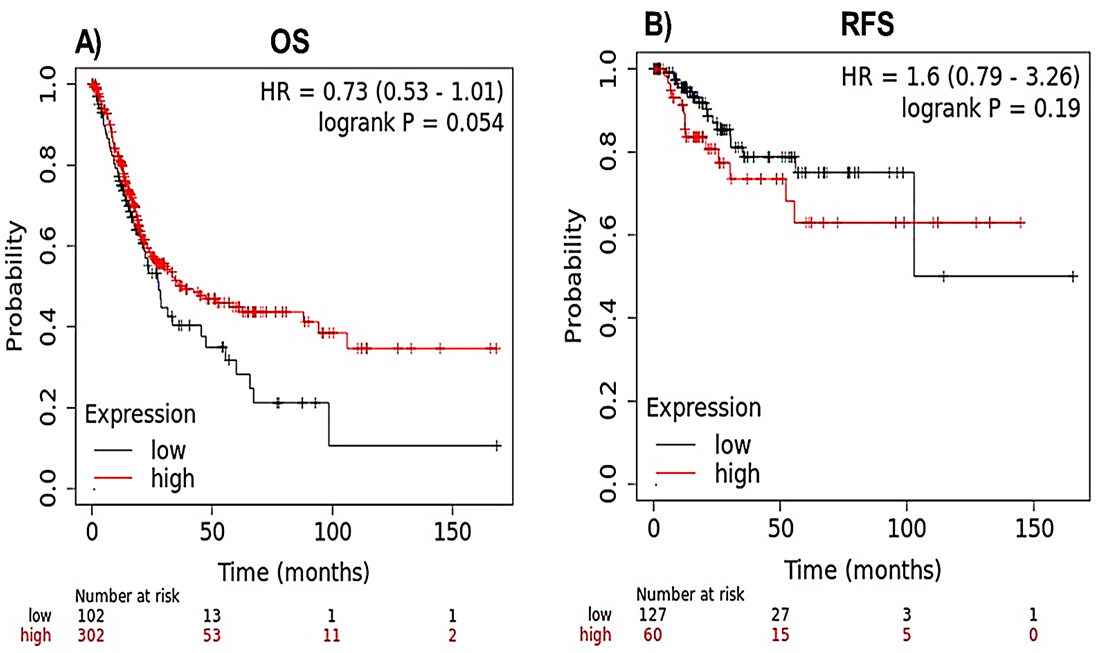
The KM plotter was applied to examine the impact of IQGAP3 expression on the patients’ survival under two “OS” and “RFS” models. However, there was no apparent connection between IQGAP3 transcription level with OS (HR = o.73 [0.53-1.01], P = 0.054) and RFS (HR = 1.6 [0.79-3.26], P = 0.19) in BLCA patients (Fig. 12).
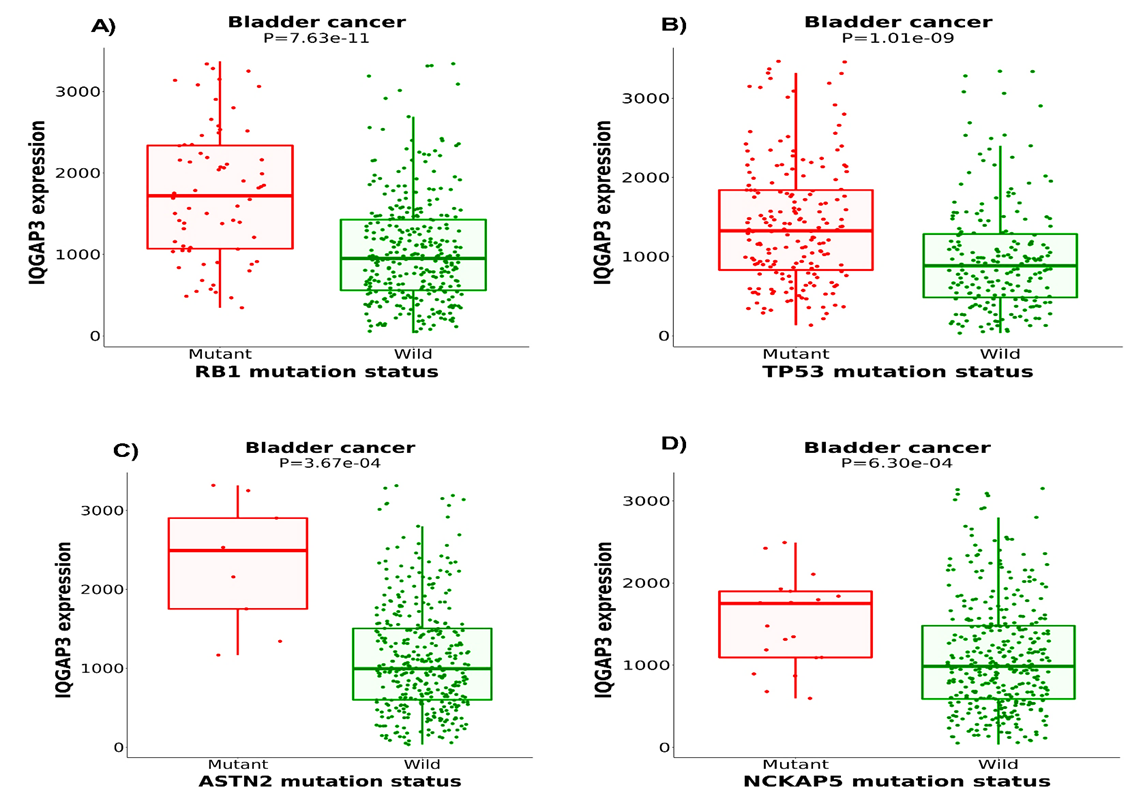
Fig. 8. The top four mutant genes positively related to IQGAP3 in BLCA from MuTarget.
IQGAP3= Isoleucine–glutamine motif-containing GTPase-activating protein 3; BLCA= Bladder urothelial carcinoma.
Fig. 9. Relationship between IQGAP3 expression and the levels of immune checkpoint inhibitors. Plots showing the correlation between IQGAP3 expression and the levels of A) PD-L1 (CD274), B) PD-1 (PDCD1), C) LAG3, and D) CTLA4 in BLCA using TIMER2 platform.
IQGAP3= Isoleucine–glutamine motif-containing GTPase-activating protein 3; PL-L1= Programmed death-ligand 1; PD-1= Programmed death protein 1; LAG3= Lymphocyte-activation gene 3; CTLA4= Cytotoxic T-lymphocyte antigen 4.
Discussion
The oncogene IQGAP3 is noticeably upregulated in a spectrum of human malignancies, including BLCA [6]. However, its role in the tumorigenesis of BLCA has been partially specified. The current study is the first multi-omics analysis to disclose the relation between IQGAP3 and BLCA. Earlier clinical observations manifested higher expression of the IQGAP3 gene in BLCA relative to normal bladder specimens. Li et al. clarified that IQGAP3 mRNA expression in BLCA was much higher than in control tissues. The protein levels of IQGAP3 assessed by immunohistochemistry staining exhibited a similar trend to their mRNA levels. Moreover, they reported that the transcription levels of IQGAP1 and IQGAP2 in BLCA cells did not show any difference from their expression in normal cells. However, IQGAP3 mRNA level in BLCA cell lines was substantially overexpressed compared to that in normal cell lines [9]. Kim et al. demonstrated that IQGAP3 mRNA levels were meaningfully raised in BLCA tissues and urine samples relative to their corresponding standard samples. That increased IQGAP3 could be a valuable diagnostic marker for discriminating BLCA cases from non-cancerous cases [10]. In agreement with these observations, we employed multiple web-based databases. We confirmed that IQGAP3 was substantially boosted in BLCA versus the typical cases, a common characteristic of oncogenic genes. Tumor stage and grade are indispensable indicators for predicting the clinical behavior of tumors and choosing the most relevant therapies [20]. Therefore, exploring the consequences of IQGAP3 upregulation on tumor stage and grade was important.
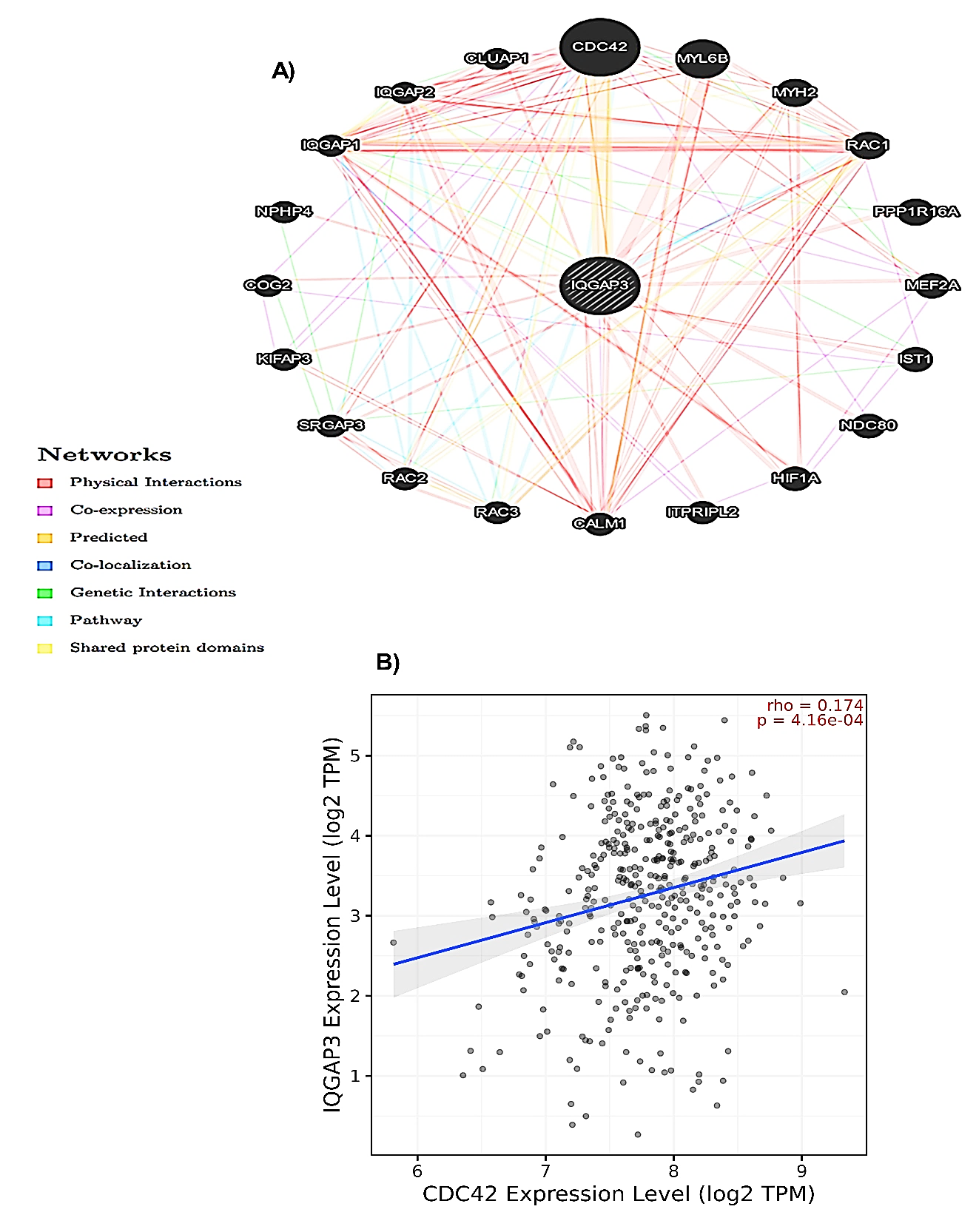
Fig. 10. The PPI network of IQGAP3 protein was constructed using GeneMANIA. A) The PPI network of IQGAP3, and B) Relationship between IQGAP3 expression and CDC42 according to the TIMER2 database. IQGAP3= Isoleucine–glutamine motif-containing GTPase-activating protein 3; PPI= Protein-protein interaction; CDC42= Cell division cycle 42.
Fig. 11. Enrichment analysis of IQGAP3 and top 50 related genes in BLCA. A) Biological processes, B) cellular components, C) Biological processes, and D) KEGG enrichment scatter plots were attained from the Enrichr database.
BLCA= Bladder urothelial carcinoma; IQGAP3= Isoleucine–glutamine motif-containing GTPase-activating protein 3; KEGG= Kyoto Encyclopedia of Genes and Genomes.
Fig. 12. Prognostic analysis of IQGAP3 in BLCA patients. A) Overall survival (OS), and B) Recurrence-free survival (RFS).
BLCA= Bladder urothelial carcinoma; IQGAP3= Isoleucine–glutamine motif-containing GTPase-activating protein 3.
A positive correlation was observed between IQGAP3 expression with tumor grade and tumor stage, indicating that IQGAP3 may play an oncogenic role and serve as a promising and novel diagnostic factor for BLCA. Subsequently, we scrutinized the potential mechanisms for IQGAP3 upregulation in BLCA. Epigenetic modifications such as DNA methylation have been associated with the regulation of gene expression. The aberrant methylation pattern in the promotor region is the most recognized epigenetic event to occur in human cancers. Hence, hypermethylation of tumor suppressor genes and hypomethylation of oncogenes are importantly linked to the neoplastic processes [21]. Our in-silico data could not establish the possibility of epigenetic regulation of IQGAP3 expression, as we did not observe a meaningful relation in the IQGAP3 promoter methylation among BLCA and non-cancerous individuals. Accordingly, another in-silico investigation based on TCGA data by Kumar et al. also clarified that increased levels of IQGAP3 in cancers were independent of hypomethylation of the IQGAP3 promoter [22]. Therefore, another complex mechanism may be involved in IQGAP3 gene regulation.
A large number of scientific studies have convincingly revealed that genetic events can outstandingly contribute to cancer initiation and progression [23]. However, little is known regarding the role of IQGAP3 genetic modifications in modulating the expression of IQGAP3 in BLCA. Our analysis of the data acquired from cBioPortal exhibited that the genetic alterations of the IQGAP3 gene occur in 25 (6%) of 411 analyzed BLCA cases. Furthermore, most alteration was related to high gene amplification, which further revealed a positive relation with the observed increased expression levels of IQGAP3 in our in-silico study. Therefore, the elevated amplification of IQGAP3 in BLCA potentiates the probability that genomic modifications in the IQGAP3 gene may be responsible, at least partly, for the IQGAP3 upregulation in BLCA.
Furthermore, we applied the muTarget platform to represent the most critical mutant genes associated with IQGAP3 expression. Among them, the most significant genes were p53 and RB1. Mutations in these eminent tumor suppressor genes have been exceedingly related to the occurrence and development of various malignancies, including BLCA [24]. Due to their critical roles in cell cycle regulation, mutations in these genes lead to the growth of tumors with high grade and invasiveness capability [25]. The analyses from the muTarget database demonstrated IQGAP3's high expression level in the BLCA samples containing mutations in p53 and RB1 genes. IQGAP3 is a suitable target for the progression of BLCA in future studies.
Previous experimental studies and our PPI data propounded that interaction between IQGAP3 and the Rho family may play a crucial role in the initiation and progression of BLCA. Rho GTPase is identified as an oncogene in different human malignancies; specifically, it exerts its impression in tumorigenesis by modulating cytoskeleton dynamics and adhesion transition [26]. As a vital member of the Rho GTPase family, activation of CDC42 leads to biological behaviors such as cytoskeletal changes, cell adhesion, proliferation, and metastasis in malignant cells [27]. IQGAP3 seems to bind to active CDC42 selectively. In a prior study, Li et al. reported that CDC42 and IQGAP3 were upregulated in BLCA cells. Later, they knocked down CDC42 and observed that cell proliferation was significantly reduced, coupled with a sharp decrease in IQGAP3, Ras, and p-Erk levels in BLCA cells. Their findings suggested that CDC42 inhibited cell proliferation in BLCA via the IQGAP3‑mediated Ras/ERK pathway [9]. Our functional analysis revealed that IQGAP3 and its correlated genes may also be involved in the cell cycle, p53 signaling pathway, mitotic nuclear division, and the oncogenic miRNAs, contributing to BLCA progression. The tumor immune microenvironment is a complex network characterized by the infiltration of immune cells with both tumor-promoting and anti-tumoral functions [28]. CD8+ T cells exert great cytotoxic activities, but these cells become exhausted under chronic stimulation situations in the tumor immune microenvironment [29]. A hallmark of CD8 exhaustion is the expression of immune checkpoints, such as PD-1 and PD-L1, within the tumor immune microenvironment that functions as a brake to impede activated CD8+ cytotoxic functions [30]. PD-L1 binds to PD-1 on activated T cells and counteract T cell-activating signals, hindering anti-tumor immunity [31]. Although immune checkpoint blockade-based therapies have been approved for BLCA patients, only a minority respond to these therapies [32]. Additional immune checkpoints may mediate resistance to cancer immunotherapy. Co-expression of PD-1 and LAG3, which is linked to T cell exhaustion, has been observed in intra-tumoral T cells [33]. The current study showed a positive relation between IQGAP3 expression with LAG3 and PD-L1, vital CD8+ T cell exhaustion indicators.
As a result, regarding the totality of the results presented in this in-silico study, it is intelligible that high IQGAP3 expression is related to BLCA development. However, we focused on bioinformatics analysis, and no clinical data and biomedical experiments were implemented to understand the in-depth mechanism of IQGAP3 in BLCA initiation and progression.
Conclusions
IQGAP3 was upregulated in BLCA, and its upregulation was positively correlated with tumor stage and grade, indicating that IQGAP3 may serve as a promising diagnostic factor for BLCA. Through the molecular and functional analyses, we elucidated a mechanistic model for the oncogenic roles of IQGAP3 in BLCA. IQGAP3 may be related to the cell cycle, p53 signaling pathway, mitotic nuclear division, and the oncogenic miRNAs. We also elucidated that IQGAP3 transcription level was related to immune modulators in BLCA. Our findings revealed that upregulation of IQGAP3 expression promoted BLCA progression. Our study suggests future in vitro and in vivo studies to ascertain the exact molecular mechanisms of IQGAP3 in BLCA.
Ethical Consideration
The ethics committee of Shahid Sadoughi University of Medical Sciences, Yazd, Iran, approved the current study (Ethics Code: IR.SSU.MEDICINE.REC.1400.358).
Funding
The present study did not receive any dedicated financial support from public, commercial, or not-for-profit funding organizations.
Conflict of Interest
The authors declare that there is no conflict of interest associated with this work.
Acknowledgments
We would like to thank the Clinical Biochemistry Department of Shahid Sadoughi University of Medical Sciences for its support.
Authors' Contributions
The study design and interpretation were conducted by Omid Abazari, Serajoddin Vahidi, Mohammad Hossein Modarressi, and Javad Zavar Reza. Omid Abazari and Sahar Valizadeh collected the data and prepared the initial draft. Serajoddin Vahidi, Mohammad Hossein Modarressi, and Javad Zavar Reza provided editing, supervision, and review. All authors approved the final version.
References
[1]. Siegel RL, Miller KD, Wagle NS, Jemal A. Cancer statistics, 2023. Cancer Journal for Clinicians 2023; 73(1): 17-48.
[2]. Sung H, Ferlay J, Siegel RL, Laversanne M, Soerjomataram I, Jemal A, et al. Global cancer statistics 2020: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin. 2021; 71(3): 209-249.
[3]. Michael J, Matulewicz RS, Bjurlin MA. Assessment of tobacco screening and smoking cessation recommendations among bladder cancer guidelines: a call to action. The Journal of Urology 2022; 207(3): 490-92.
[4]. Liu Z, Liu X, Liu F, Zhao H, Zhang Y, Wang Y, et al. The comprehensive and systematic identification of BLCA-specific SF-regulated, survival-related AS events. Gene. 2022; 835: 146657.
[5]. van Hoogstraten LMC, Vrieling A, van der Heijden AG, Kogevinas M, Richters A, Kiemeney LA. Global trends in the epidemiology of bladder cancer: challenges for public health and clinical practice. Nat Rev Clin Oncol. 2023; 20(5): 287-304.
[6]. Song F, Dai Q, Grimm MO, Steinbach D. The antithetic roles of IQGAP2 and IQGAP3 in cancers. Cancers 2023; 15(4): 1115.
[7]. Song F, Kotolloshi R, Gajda M, Hölzer M, Grimm MO, Steinbach D. Reduced IQGAP2 promotes bladder cancer through regulation of MAPK/ERK pathway and cytokines. Int J Mol Sci. 2022; 23(21): 13508.
[8]. Mei W, Dong Y, Gu Y, Kapoor A, Lin X, Su Y, et al. IQGAP3 is relevant to prostate cancer: A detailed presentation of potential pathomechanisms. J Adv Res. 2023; 54: 195-210.
[9]. Li G, Wang Y, Guo XB, Zhao B. CDC42 regulates cell proliferation and apoptosis in bladder cancer via the IQGAP3-mediated ras/ERK pathway. Biochem Genet. 2022; 60(6): 2383-98.
[10]. Kim WT, Kim YH, Jeong P, Seo SP, Kang HW, Kim YJ, et al. Urinary cell-free nucleic acid IQGAP3: A new non-invasive diagnostic marker for bladder cancer. Oncotarget. 2018; 9(18): 14354.
[11]. Bartha Á, Győrffy B. TNMplot. com: A web tool for the comparison of gene expression in normal, tumor and metastatic tissues. Int J Mol Sci. 2021; 22(5): 2622.
[12]. Chandrashekar DS, Karthikeyan SK, Korla PK, Patel H, Shovon AR, Athar M, et al. UALCAN: An update to the integrated cancer data analysis platform. Neoplasia. 2022; 25: 18-27.
[13]. Goldman MJ, Craft B, Hastie M, Repečka K, McDade F, Kamath A, et al. Visualizing and interpreting cancer genomics data via the Xena platform. Nat Biotechnol. 2020; 38(6): 675-78.
[14]. Győrffy B, Surowiak P, Budczies J, Lánczky A. Online survival analysis software to assess the prognostic value of biomarkers using transcriptomic data in non-small-cell lung cancer. PloS one. 2013; 8(12): e82241.
[15]. Franz M, Rodriguez H, Lopes C, Zuberi K, Montojo J, Bader GD, et al. GeneMANIA update 2018. Nucleic Acids Res. 2018; 46(1): 60-4.
[16]. Cerami E, Gao J, Dogrusoz U, Gross BE, Sumer SO, Aksoy BA, et al. The cBio cancer genomics portal: an open platform for exploring multidimensional cancer genomics data. Cancer Discov. 2012; 2(5): 401-404.
[17]. Xie Z, Bailey A, Kuleshov M V, Clarke DJB, Evangelista JE, Jenkins SL, et al. Gene set knowledge discovery with Enrichr. Curr Protoc. 2021; 1(3): 90.
[18]. Nagy Á, Győrffy B. muTarget: a platform linking gene expression changes and mutation status in solid tumors. Int J cancer. 2021; 148(2): 502-11.
[19]. Li T, Fan J, Wang B, Traugh N, Chen Q, Liu JS, et al. TIMER: a web server for comprehensive analysis of tumor-infiltrating immune cells. Cancer Res. 2017; 77(21): e108-10.
[20]. Telloni SM. Tumor staging and grading: A primer. Mol Profiling Methods Protoc. 2017; 1-17.
[21]. Ibrahim J, Peeters M, Van Camp G, de Beeck KO. Methylation biomarkers for early cancer detection and diagnosis: Current and future perspectives. Eur J Cancer. 2023; 178: 91-113.
[22]. Kumar D, Hassan MK, Pattnaik N, Mohapatra N, Dixit M. Reduced expression of IQGAP2 and higher expression of IQGAP3 correlates with poor prognosis in cancers. PLoS One 2017; 12(10): 186977.
[23]. Schulz WA. An introduction to human cancers. Mol Biol Hum Cancers. 2023; 3(1): 3-28.
[24]. Hurst CD, Knowles MA. Mutational landscape of non-muscle-invasive bladder cancer. In: Urologic Oncology: Seminars and Original Investigations. Elsevier 2022; 40(7): 295-303.
[25]. Cooley LF, Glaser AP, Meeks JJ. Mutation signatures to Pan-Cancer Atlas: Investigation of the genomic landscape of muscle-invasive bladder cancer. In: Urologic Oncology: Seminars and Original Investigations. Elsevier 2022; 40(7): 279-86.
[26]. Crosas-Molist E, Samain R, Kohlhammer L, Orgaz JL, George SL, Maiques O, et al. Rho GTPase signaling in cancer progression and dissemination. Physiol Rev. 2022; 102(1): 455-510.
[27]. Bailly C, Beignet J, Loirand G, Sauzeau V. Rac1 as a therapeutic anticancer target: Promises and limitations. Biochem Pharmacol. 2022; 203: 115180.
[28]. Jia Q, Wang A, Yuan Y, Zhu B, Long H. Heterogeneity of the tumor immune microenvironment and its clinical relevance. Exp Hematol Oncol. 2022; 11(1): 24-30.
[29]. Huang Y, Jia A, Wang Y, Liu G. CD8+ T cell exhaustion in anti‐tumour immunity: The new insights for cancer immunotherapy. Immunology. 2023; 168(1): 30-48.
[30]. Zhang L, Zhang B, Li L, Ye Y, Wu Y, Yuan Q, et al. Novel targets for immunotherapy associated with exhausted CD8+ T cells in cancer. J Cancer Res Clin Oncol. 2023; 149(5): 2243-258.
[31]. Pei L, Liu Y, Liu L, Gao S, Gao X, Feng Y, et al. Roles of cancer-associated fibroblasts (CAFs) in anti-PD-1/PD-L1 immunotherapy for solid cancers. Mol Cancer. 2023; 22(1): 29-35.
[32]. Liu Z, Zeng H, Jin K, Yu Y, You R, Zhang H, et al. TIGIT and PD-1 expression atlas predicts response to adjuvant chemotherapy and PD-L1 blockade in muscle-invasive bladder cancer. Br J Cancer. 2022; 126(9): 1310-317.
[33]. Jiang H, Ni H, Zhang P, Guo X, Wu M, Shen H, et al. PD-L1/LAG-3 bispecific antibody enhances tumor-specific immunity. Oncoimmunology 2021; 10(1): 1943180.










































.jpg)