Fri, Feb 13, 2026
[Archive]
Volume 8, Issue 3 (August 2021)
IJML 2021, 8(3): 180-187 |
Back to browse issues page
Download citation:
BibTeX | RIS | EndNote | Medlars | ProCite | Reference Manager | RefWorks
Send citation to:



BibTeX | RIS | EndNote | Medlars | ProCite | Reference Manager | RefWorks
Send citation to:
Zolfaghari M R. Detection of BlaVIM and BlaIMP Carbapenemase-Producing Pseudomonas Aeruginosa from Clinical Samples in Iran. IJML 2021; 8 (3) :180-187
URL: http://ijml.ssu.ac.ir/article-1-393-en.html
URL: http://ijml.ssu.ac.ir/article-1-393-en.html
Department of Biology, Qom Branch, Islamic Azad University, Qom, Iran
Full-Text [PDF 269 kb]
(619 Downloads)
| Abstract (HTML) (1390 Views)
References
Full-Text: (557 Views)
Introduction
Pseudomonadsae aeruginosa (P. aeruginosa) is a gram-negative, aerobic, rod-shaped bacterium. Moreover, P. aeruginosa is an opportunistic pathogen causing many infections such as urinary tract infections (UTI), respiratory infections, wounds, and nosocomial infections in humans [1]. Beta-lactam antibiotics are the first drugs used to treat these infections, among which carbapenems are the most effective ones against P. aeruginosa that can be used to treat infections caused by this bacterium. However, unfortunately, we are currently witnessing widespread resistance to various types of beta-lactam antibiotics worldwide [2-4].
The mechanism of bacterial resistance to antibiotics is very diverse. Mechanisms involved in developing resistance to carbapenems are integrin or plasmid-mediated carbapenemases, the reduced porin expression, the increased chromosomal cephalosporins activity, and the increased expression of efflux systems [5]. The common carbapenemases in P. aeruginosa are mainly Metallo-β-lactamases of Verona integron-encoded Metallo-β-lactamase, active on imipenem (IMP), São Paulo metalo-beta-lactamase, German imipenemase, Adelaide imipenemase, Dutch imipenemase, and New Delhi metallo-beta-lactamase types. Accordingly, IMP was firstly identified in Japan, and VIM was found in Italy. However, recently, the prevalence of these genes is reported worldwide [6-9].
The presence of carbapenemase genes on plasmids transfers them between different species and bacterial genera [7]. The accurate detection of carbapenemases in bacteria is not easy. Although the Modified Hodge Test (MHT) is a relatively easy and simple phenotypic test, it is not recommended due to its low accuracy and sensitivity. The identification of carbapenemase genes using molecular methods, e.g., polymerase chain reaction (PCR), is usually reliable and accurate; however, it requires specialized knowledge in the epidemiology of carbapenemases [10].
This study aimed to detect blaIMP and blaVIM carbapenemases by MHT and reverse transcription-polymerase chani reaction (RT-PCR) among P. aeruginosa isolated in some hospitals in Iran.
Materials and Methods
Sample collection
In this cross-sectional study, P. aeruginosa strains were collected from different clinical samples such as burn, blood, wound, and other liquids body in Firoozgar and Shahid Motahari Hospitals in Tehran and Velayat Hospital Rasht Province, from May to December 2018. The initial identification was performed based on conventional bacteriological and biochemical methods such as colony morphology, oxidase, catalase, Indol, Motility, citrate, and triple sugar iron [11].
Antibiotic resistance pattern
The antibiotic susceptibility test was determined using the Kirby-Bauer method in terms of the Clinical Laboratory Standard Institute guidelines (CLSI 2017). The antibiotic discs use as follows: ceftazidime (30 μg), cefotaxime (30 μg), cefepime (30 μg), ceftriaxone (30 μg), imipenem (10 µg), meropenem (10 μg), ertapenem (10 µg), gentamicin (10 μg), piperacillin (100 μg), and aztreonam (30 μg) (MAST Company, UK) [12]. P. aeruginosa ATCC 27853 was used as the positive control strain in the antimicrobial susceptibility test [13].
MHT for the detection of carbapenemase
The MHT was performed according to CLSI using E. coli ATCC 25922 and meropenem at the dose of 10 μg (Oxoid, UK). In order to perform this test, E. coli ATCC 25922 was cultured on Mueller-Hinton agar, and 0.5 McFarland turbidity of P. aeruginosa isolates were then inoculated on the surfaces of plates. Afterward, a disc of meropenem at 10 μg (Oxoid, UK) was placed in the center. After the incubation for 18 hours at 370C, a positive result was observed by the enhanced growth of the indicator strain towards a meropenem disc, clover leaf-type indentation at the point of intersection of the isolate with the indicator strain [14]. P. aeruginosa ATCC 15442 and P. aeruginosa ATCC 27853 were used as negative and positive controls, respectively.
Detection of blaIMP and blaVIM genes
According to the manufacturer's guidelines, total RNA isolation was done, and DNA was removed using 20 U of RQ1 DNase I (Sinaclon, Iran). After that, cDNA was synthesized by Pars Tous kit and then stored at -20 ºC.
In this study, RT-PCR was used due to time- and cost-effectiveness and less error due to less contamination. BlaIMP and blaVIM were detected in all the isolates by special primers designed via IDT software and then synthesized by Macrogen Company (Korea) (Table 1). Finally, PCR was performed on the RT reaction product [15]. Positive control isolates were obtained from Rouhi et al., who studied Metallo-beta-lactamase genes in Kurdistan Province, Iran [16]. This research was approved by the Ethics Committee of Islamic Azad University, Qom, Iran.
Statistical analysis
T-test and Pearson’s chi-square test were used for data analyses by SPSS version 23.0 (SPSS, Chicago, IL). The level of significance in the current study was considered as <0.05.
Results
One hundred P. aeruginosa were isolated in Tehran. These isolates had the following properties: Gram-negative bacilli, oxidase-positive, catalase-positive, Indol negative, motile, citrate positive, and triple sugar iron Alk/Alk. In addition, the specimens included burn (N = 55, 55%), blood (N = 2, 2%), wound (N = 16, 16%), and other liquids body (N = 27, 27%). Resistance rates of all the clinical isolates against the tested antibiotics were as follows: ceftazidime 39%, cefotaxime 51%, cefepime 28%, ceftriaxone 34%,imipenem 46%, meropenem 32%, ertapenem 41%, gentamicin 41%, piperacillin 35%, and aztreonam 30%. Therefore, the highest and lowest resistance rates were related to cefotaxime and cefepime, respectively. The results of the antibiotic resistance pattern of P. aeruginosa isolates in the different clinical specimens of this study are shown in Table 2. In this study, those semi-sensitive isolates were considered resistant isolates. To note, the burn samples had the highest resistance to ertapenem and the lowest resistance to piperacillin. Among the wound samples, the highest and lowest rates of antibiotic resistance were related to imipenem and ertapenem, respectively. However, among P. aeruginosa strains isolated from other body fluids, the highest and lowest resistance rates were related to cefotaxime and gentamicin, respectively.
The mechanism of bacterial resistance to antibiotics is very diverse. Mechanisms involved in developing resistance to carbapenems are integrin or plasmid-mediated carbapenemases, the reduced porin expression, the increased chromosomal cephalosporins activity, and the increased expression of efflux systems [5]. The common carbapenemases in P. aeruginosa are mainly Metallo-β-lactamases of Verona integron-encoded Metallo-β-lactamase, active on imipenem (IMP), São Paulo metalo-beta-lactamase, German imipenemase, Adelaide imipenemase, Dutch imipenemase, and New Delhi metallo-beta-lactamase types. Accordingly, IMP was firstly identified in Japan, and VIM was found in Italy. However, recently, the prevalence of these genes is reported worldwide [6-9].
The presence of carbapenemase genes on plasmids transfers them between different species and bacterial genera [7]. The accurate detection of carbapenemases in bacteria is not easy. Although the Modified Hodge Test (MHT) is a relatively easy and simple phenotypic test, it is not recommended due to its low accuracy and sensitivity. The identification of carbapenemase genes using molecular methods, e.g., polymerase chain reaction (PCR), is usually reliable and accurate; however, it requires specialized knowledge in the epidemiology of carbapenemases [10].
This study aimed to detect blaIMP and blaVIM carbapenemases by MHT and reverse transcription-polymerase chani reaction (RT-PCR) among P. aeruginosa isolated in some hospitals in Iran.
Materials and Methods
Sample collection
In this cross-sectional study, P. aeruginosa strains were collected from different clinical samples such as burn, blood, wound, and other liquids body in Firoozgar and Shahid Motahari Hospitals in Tehran and Velayat Hospital Rasht Province, from May to December 2018. The initial identification was performed based on conventional bacteriological and biochemical methods such as colony morphology, oxidase, catalase, Indol, Motility, citrate, and triple sugar iron [11].
Antibiotic resistance pattern
The antibiotic susceptibility test was determined using the Kirby-Bauer method in terms of the Clinical Laboratory Standard Institute guidelines (CLSI 2017). The antibiotic discs use as follows: ceftazidime (30 μg), cefotaxime (30 μg), cefepime (30 μg), ceftriaxone (30 μg), imipenem (10 µg), meropenem (10 μg), ertapenem (10 µg), gentamicin (10 μg), piperacillin (100 μg), and aztreonam (30 μg) (MAST Company, UK) [12]. P. aeruginosa ATCC 27853 was used as the positive control strain in the antimicrobial susceptibility test [13].
MHT for the detection of carbapenemase
The MHT was performed according to CLSI using E. coli ATCC 25922 and meropenem at the dose of 10 μg (Oxoid, UK). In order to perform this test, E. coli ATCC 25922 was cultured on Mueller-Hinton agar, and 0.5 McFarland turbidity of P. aeruginosa isolates were then inoculated on the surfaces of plates. Afterward, a disc of meropenem at 10 μg (Oxoid, UK) was placed in the center. After the incubation for 18 hours at 370C, a positive result was observed by the enhanced growth of the indicator strain towards a meropenem disc, clover leaf-type indentation at the point of intersection of the isolate with the indicator strain [14]. P. aeruginosa ATCC 15442 and P. aeruginosa ATCC 27853 were used as negative and positive controls, respectively.
Detection of blaIMP and blaVIM genes
According to the manufacturer's guidelines, total RNA isolation was done, and DNA was removed using 20 U of RQ1 DNase I (Sinaclon, Iran). After that, cDNA was synthesized by Pars Tous kit and then stored at -20 ºC.
In this study, RT-PCR was used due to time- and cost-effectiveness and less error due to less contamination. BlaIMP and blaVIM were detected in all the isolates by special primers designed via IDT software and then synthesized by Macrogen Company (Korea) (Table 1). Finally, PCR was performed on the RT reaction product [15]. Positive control isolates were obtained from Rouhi et al., who studied Metallo-beta-lactamase genes in Kurdistan Province, Iran [16]. This research was approved by the Ethics Committee of Islamic Azad University, Qom, Iran.
Statistical analysis
T-test and Pearson’s chi-square test were used for data analyses by SPSS version 23.0 (SPSS, Chicago, IL). The level of significance in the current study was considered as <0.05.
Results
One hundred P. aeruginosa were isolated in Tehran. These isolates had the following properties: Gram-negative bacilli, oxidase-positive, catalase-positive, Indol negative, motile, citrate positive, and triple sugar iron Alk/Alk. In addition, the specimens included burn (N = 55, 55%), blood (N = 2, 2%), wound (N = 16, 16%), and other liquids body (N = 27, 27%). Resistance rates of all the clinical isolates against the tested antibiotics were as follows: ceftazidime 39%, cefotaxime 51%, cefepime 28%, ceftriaxone 34%,imipenem 46%, meropenem 32%, ertapenem 41%, gentamicin 41%, piperacillin 35%, and aztreonam 30%. Therefore, the highest and lowest resistance rates were related to cefotaxime and cefepime, respectively. The results of the antibiotic resistance pattern of P. aeruginosa isolates in the different clinical specimens of this study are shown in Table 2. In this study, those semi-sensitive isolates were considered resistant isolates. To note, the burn samples had the highest resistance to ertapenem and the lowest resistance to piperacillin. Among the wound samples, the highest and lowest rates of antibiotic resistance were related to imipenem and ertapenem, respectively. However, among P. aeruginosa strains isolated from other body fluids, the highest and lowest resistance rates were related to cefotaxime and gentamicin, respectively.
Table 1. Primer sequences of blaIMP and blaVIM genes and reaction setup for RT-PCR and PCR
| Primer sequences | RT-PCR | PCR | |||
| Conditions | Volume reactions | Conditions | Volume reactions | Fragment size | |
| blaIMP F: 5′-AAA GAT ACT GAA AAG TTA GT-3' R: 5' -TCY CCA AYT TCA CTR TGA CT-3' blaVIM F: 5'-CCGATGGTGTTTGGTCGCAT-3´ R: 5´-GAATGCGLAGCACCAGGAT-3´ |
1 cycle 25 ˚C 10 min 47 ˚C 60 min 70 ˚C 10 min |
Total RNA 5 μl Random hexamer 2 μl H2O up to 10 μl |
1cycle: 94 ˚C 5 min 30 cycle: 94 ˚C 30s 58 ˚C 30s 72 ˚C 30s 1cycle: 72 ˚C 8 min |
10X PCR Buffer:2.5 μl 10 mM dNTPs: 0.5 μl 10 mM MgCl2:0.75 μl 10 pmol F+R Primer: 1.25 μl Taq DNA polymerase (5u μl): 0.2 μl Template DNA: 1 μl H2O up to 25 μl |
391bp 446bp |
Table 2. Antibiotic resistance pattern in the different clinical specimens
| Antibiotics | Burn | Blood | Wound | Other liquites | ||||
| Resistance N |
Susceptible N |
Resistance N |
Susceptible N |
Resistance N |
Susceptible N |
Resistance N |
Susceptible N |
|
| Ceftazidime | 23 | 32 | 1 | 1 | 6 | 10 | 9 | 18 |
| Cefotaxime | 20 | 35 | 2 | 0 | 11 | 5 | 18 | 9 |
| Cefepime | 13 | 42 | 0 | 2 | 5 | 11 | 10 | 17 |
| Ceftriaxone | 16 | 39 | 1 | 1 | 4 | 12 | 13 | 14 |
| Imipenem | 18 | 37 | 1 | 1 | 12 | 4 | 15 | 12 |
| Meropenem | 18 | 37 | 0 | 2 | 5 | 11 | 9 | 18 |
| Ertapenem | 29 | 26 | 1 | 1 | 4 | 12 | 7 | 20 |
| Gentamicin | 31 | 24 | 0 | 2 | 6 | 10 | 4 | 23 |
| Piperacillin | 12 | 43 | 1 | 1 | 5 | 11 | 17 | 10 |
| Aztreonam | 8 | 47 | 1 | 1 | 8 | 8 | 13 | 14 |
Table 3. Distribution of carbapenemase genes in the different clinical specimens
| Source of sample | ||||||
| Total | Other liquids | Wound | Blood | Burn | Gene (s) | |
| 83 | 11 | 16 | 1 | 55 | blaVIM | |
| 11 | 2 | 2 | 0 | 7 | blaIMP | |
| 11 | 2 | 2 | 0 | 7 | blaVIM/ blaIMP | |

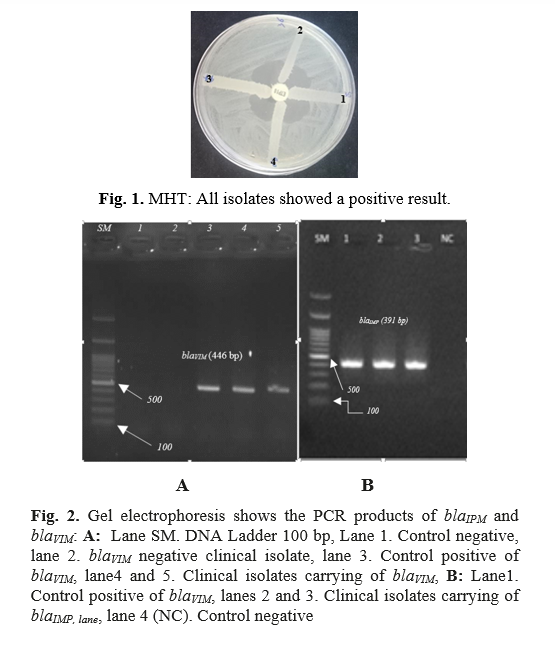
The result of MHT showed that 74 isolates (74%) were positive in this research. In addition, the results of the standard reference MIC tests were in fair agreement with those of the MHT (N = 74) (Figure 1). BlaVIM and blaIMP genes were detected in 83% and 11% of the samples, respectively (Figure 2). The results of the distribution of carbapenemase genes in the different clinical specimens are shown in Table 3.
Discussion
Recently, we observed the increasing prevalence of high antibiotic resistance in bacteria causing various infections. Since treating these infections is very difficult and has raised concerns such as the reduced treatment choices and the increased costs and mortality, it is currently considered a problem in health centers [17-19]. One of the causes of antibiotic resistance is carbapenemase enzymes, which can be easily transferred to bacteria due to being transported by integrons and then increase resistance. So, identifying these types of resistance and treating these patients as soon as possible can help control the infection [19]. Therefore, it was attempted to isolate these isolates in the studied hospitals using an accurate and reliable molecular technique in this study.
Studying the frequency of antibiotic resistance of the tested isolates showed that the resistance of the strains against therapeutic antibiotics is high. The highest resistance rate was found to be related to cefotaxime (51%). Correspondingly, in another study conducted by Bagheri Bejestani et al., the highest resistance rate was detected against cefotaxime (36.6%) [20]. In the current study, the resistance rate to imipenem was 46% using the disc diffusion method, similar to Saberi’s study (33 %) in Rasht [21]. However, this rate was higher than those of Bagheri Bejestani’s study (15.5%) in Iran [20] and Rouhi’s study (19.23%) in Iran [16] and lower than those of Wang’s study (77.5% ) in China [22], Abaza’s study (78.3%) in Egypt [23], and Li’s study (73.3%) in China [24]. Differences in the results of various studies may be due to different antibiotic therapy methods. In our study, 74% of the strains were phenotypically carbapenemase positive. In Othman and Shahcheraghi’s studies, 28.1% and 54% of the isolates were positive for carbapenemases by MHT, respectively [25, 26]. MHT is an easy, cost-effective, and fast method to identify carbapenemase strains. The accuracy and sensitivity of this test are less than molecular techniques, but due to its high-speed capability, it is recommended to identify and isolate more carbapenemase-producing isolates.
The difference among the frequencies of carbapenemase-producing isolates by the MHT phenotypic method in different studies may be due to regional differences and various patterns of antibiotic use [27].
The result of the RT-PCR method related to blaIMP and blaVIM genes indicated that 83% of P. aeruginosa strains harbored the blaVIM gene, and 11% of them carried blaIMP gene. Other studies performed in Portugal and China confirmed 19.4% and 43.5% of blaVIM gene in P. aeruginosa strains, respectively [28, 29]. Additionally, the rate of blaIMP gene in the tested isolates was similar to those of the Cheng et al. and Othman et al.’s studies (26, 29). Alkhudhairy in Iraq reported 33.3% and 25% strains carrying blaVIM and blaIMP genes, respectively [30].
Differences in the results of different studies can be due to differences in time, geographical area, and even the test methods used. The pattern of antibiotic use varies at different times and geographical locations, which can affect the prevalence of carbapenemase genes. Besides, the differences in the results related to the frequency of genes can be due to the increasing resistance to cephalosporins of the third generation, thereby more carbapenems consumption in recent years [31].
According to previous reports performed in this regard, the blaVIM gene in P. aeruginosa strains is more abundant than other Metallo-beta-lactamase genes, which also is the dominant Metallo-beta-lactamase gene in Iran [25, 32].
In the current study, the isolates that did not carry the genes tested, but resulted as positive for carbapenemase using phenotype test, were likely to carry other carbapenemase genes that were not tested due to the cost and time required to perform these tests.
Conclusions
As the blaVIM and blaIMP genes are carried by integrons, which can carry other antibiotic resistance genes, they make it difficult to treat infections resulted from these strains. Therefore, the detection of strains carrying these genes can be an effective step in treating these infections in different rejoins.
Consequently, performing phenotypic tests to evaluate antibiotic susceptibility and resistance before antibiotic administration is unavoidable. Moreover, molecularly studying carbapenemase-producing enzyme genes due to their wide variety and prevalence rates in different geographical areas to control this strain is essential.
Conflicts of Interest
There are no conflicts of interest.
Acknowledgments
The authors would like to thank Dr. Hossein Keyvani, Iran University of Medical Sciences, who contributed to this study.
Discussion
Recently, we observed the increasing prevalence of high antibiotic resistance in bacteria causing various infections. Since treating these infections is very difficult and has raised concerns such as the reduced treatment choices and the increased costs and mortality, it is currently considered a problem in health centers [17-19]. One of the causes of antibiotic resistance is carbapenemase enzymes, which can be easily transferred to bacteria due to being transported by integrons and then increase resistance. So, identifying these types of resistance and treating these patients as soon as possible can help control the infection [19]. Therefore, it was attempted to isolate these isolates in the studied hospitals using an accurate and reliable molecular technique in this study.
Studying the frequency of antibiotic resistance of the tested isolates showed that the resistance of the strains against therapeutic antibiotics is high. The highest resistance rate was found to be related to cefotaxime (51%). Correspondingly, in another study conducted by Bagheri Bejestani et al., the highest resistance rate was detected against cefotaxime (36.6%) [20]. In the current study, the resistance rate to imipenem was 46% using the disc diffusion method, similar to Saberi’s study (33 %) in Rasht [21]. However, this rate was higher than those of Bagheri Bejestani’s study (15.5%) in Iran [20] and Rouhi’s study (19.23%) in Iran [16] and lower than those of Wang’s study (77.5% ) in China [22], Abaza’s study (78.3%) in Egypt [23], and Li’s study (73.3%) in China [24]. Differences in the results of various studies may be due to different antibiotic therapy methods. In our study, 74% of the strains were phenotypically carbapenemase positive. In Othman and Shahcheraghi’s studies, 28.1% and 54% of the isolates were positive for carbapenemases by MHT, respectively [25, 26]. MHT is an easy, cost-effective, and fast method to identify carbapenemase strains. The accuracy and sensitivity of this test are less than molecular techniques, but due to its high-speed capability, it is recommended to identify and isolate more carbapenemase-producing isolates.
The difference among the frequencies of carbapenemase-producing isolates by the MHT phenotypic method in different studies may be due to regional differences and various patterns of antibiotic use [27].
The result of the RT-PCR method related to blaIMP and blaVIM genes indicated that 83% of P. aeruginosa strains harbored the blaVIM gene, and 11% of them carried blaIMP gene. Other studies performed in Portugal and China confirmed 19.4% and 43.5% of blaVIM gene in P. aeruginosa strains, respectively [28, 29]. Additionally, the rate of blaIMP gene in the tested isolates was similar to those of the Cheng et al. and Othman et al.’s studies (26, 29). Alkhudhairy in Iraq reported 33.3% and 25% strains carrying blaVIM and blaIMP genes, respectively [30].
Differences in the results of different studies can be due to differences in time, geographical area, and even the test methods used. The pattern of antibiotic use varies at different times and geographical locations, which can affect the prevalence of carbapenemase genes. Besides, the differences in the results related to the frequency of genes can be due to the increasing resistance to cephalosporins of the third generation, thereby more carbapenems consumption in recent years [31].
According to previous reports performed in this regard, the blaVIM gene in P. aeruginosa strains is more abundant than other Metallo-beta-lactamase genes, which also is the dominant Metallo-beta-lactamase gene in Iran [25, 32].
In the current study, the isolates that did not carry the genes tested, but resulted as positive for carbapenemase using phenotype test, were likely to carry other carbapenemase genes that were not tested due to the cost and time required to perform these tests.
Conclusions
As the blaVIM and blaIMP genes are carried by integrons, which can carry other antibiotic resistance genes, they make it difficult to treat infections resulted from these strains. Therefore, the detection of strains carrying these genes can be an effective step in treating these infections in different rejoins.
Consequently, performing phenotypic tests to evaluate antibiotic susceptibility and resistance before antibiotic administration is unavoidable. Moreover, molecularly studying carbapenemase-producing enzyme genes due to their wide variety and prevalence rates in different geographical areas to control this strain is essential.
Conflicts of Interest
There are no conflicts of interest.
Acknowledgments
The authors would like to thank Dr. Hossein Keyvani, Iran University of Medical Sciences, who contributed to this study.
References
- Moradali MF, Ghods S, Rehm BH. Pseudomonas aeruginosa lifestyle: a paradigm for adaptation, survival, and persistence. Front Cel Infect Microbiol. 2017; 7(1): 39-47.
- Montravers P, Bassetti M. The ideal patient profile for new beta-lactam/beta-lactamase inhibitors. Curr Opinion Infect Dis. 2018; 31(6): 587-93.
- Clegg WJ, Pacilli M, Kemble SK, Kerins JL, Hassaballa A, Kallen AJ, et al. Notes from the field: large cluster of Verona integron-encoded metallo-beta-lactamase-producing carbapenem-resistant P. aeruginosa isolates colonizing residents at a skilled nursing facility-Chicago, Illinois, November 2016-March 2018. Morbid Mortal Week Rep. 2018; 67(40): 1130-138.
- Zubair K, Iregbu K. Resistance pattern and detection of metallo-beta-lactamase genes in clinical isolates of Pseudomonas aeruginosa in a central Nigeria tertiary hospital. Niger J Clinic Pract. 2018; 21(2): 176-82.
- Riera E, Cabot G, Mulet X, García-Castillo M, del Campo R, Juan C, et al. Pseudomonas aeruginosa carbapenem resistance mechanisms in Spain: impact on the activity of imipenem, meropenem and doripenem. J Antimicrob Chemother. 2011; 66(9): 2022-2027.
- Walsh TR, Toleman MA, Poirel L, Nordmann P. Metallo-β-lactamases: the quiet before the storm? Clinic Microbiol Rev. 2005; 18(2): 306-325.
- Poirel L, Walsh TR, Cuvillier V, Nordmann P. Multiplex PCR for detection of acquired carbapenemase genes. Diagnos Microbiol Infect Dis. 2011; 70(1): 119-23.
- Dortet L, Poirel L, Nordmann P. Rapid detection of carbapenemase-producing Pseudomonas spp. J Clinic Microbiol. 2012; 50(11): 3773-776.
- El-Domany RA, Emara M, El-Magd MA, Moustafa WH, Abdeltwab NM. Emergence of imipenem-resistant Pseudomonas aeruginosa clinical isolates from Egypt Coharboring VIM and IMP carbapenemases. Microb Drug Resist. 2017; 23(6): 682-86.
- Khorvash F, Yazdani M, Shabani S, Soudi A. Pseudomonas aeruginosa-producing metallo-β-lactamases (VIM, IMP, SME, and AIM) in the clinical isolates of intensive care units, a university hospital in Isfahan, Iran. Adv Biomed Res. 2017; 6(1): 1-9.
- Safari M, Alikhani MY, Arabestani MR, Kakhki RK, Jafari R. Prevalence of Metallo-β-lactamases encoding genes among pseudomonas aeruginosa strains isolated from the bedridden patients in the intensive care units. Avicenna J Clinic Microbiol Infect. 2014; 1(1): 19216-9221.
- Patel JB, ed. Performance standards for antimicrobial susceptibility testing. Wayne, PA: Clinical and laboratory standards institute; 2017.
- Sachdeva R, Sharma B, Sharma R. Evaluation of different phenotypic tests for detection of metallo-β-lactamases in imipenem-resistant Pseudomonas aeruginosa. J Lab Physician. 2017; 9(4): 249-56.
- Noyal M, Menezes G, Harish B, Sujatha S, Parija S. Simple screening tests for detection of carbapenemases in clinical isolates of nonfermentative Gram-negative bacteria. Indian J Med Res. 2009; 129(6): 350-56.
- Mohammadi Bandari N, Keyvani H, Zargar M, Talebi M, Zolfaghari MR. Epidemiological and genetic overview of the klebsiella pneumoniae carbapenemases (KPCs) in k. pneumoniae isolated from the clinical samples in Iran. International Journal of Adv Biol Biomed Res. 2018; 8(1): 75-85.
- Rouhi S, Ramazanzadeh R. Antibiotic resistance pattern and risk factors associated with urinary tract infections with pseudomonas aeruginosa among women in North West of Iran. Crescent Journal of Medical and Biological Sciences 2020; 6(4): 498-504.
- Yin S, Chen P, You B, Zhang Y, Jiang B, Huang G, et al. Molecular typing and carbapenem resistance mechanisms of Pseudomonas aeruginosa isolated from a Chinese burn center from 2011 to 2016. Front Microbiol. 2018; 9(10): 1135-141.
- Ssekatawa K, Byarugaba DK, Wampande E, Ejobi F. A systematic review: the current status of carbapenem resistance in East Africa. BMC Research Notes 2018; 11(1): 629-35.
- Nordmann P, Poirel L. Epidemiology and diagnostics of carbapenem resistance in gram-negative bacteria. Clinic Infect Dis. 2019; 69(S7): 521-28.
- Bagheri Bejestani F, Hakemi-Vala M, Momtaheni R, Bagheri Bejestani O, Gholami M. The frequency of imp and vim genes among Pseudomonas aeruginosa isolates from children’s medical center of Tehran. ArchivClinic Infect Dis. 2015; 10(1): 1-10.
- Saberi M, Zamani H, Salehzadeh A. Prevalence of IMP and VIM Metallo-Beta-Lactamases in Pseudomonas aeruginosa Isolates from Clinical and Environmental Specimens in Intensive Care Units (ICUs) of Rasht Hospitals, Iran. J Med Microbiol Infect Dis. 2015; 3(3): 62-6.
- Wang W, Wang X. Prevalence of metallo-β-lactamase genes among Pseudomonas aeruginosa isolated from various clinical samples in China. J Lab Med. 2020; 44(4): 197-203.
- Abaza AF, El Shazly SA, Selim H, Aly G. Metallo-beta-lactamase producing Pseudomonas aeruginosa in a healthcare setting in Alexandria, Egypt. Pol J Microbiol. 2017; 66(3): 297-308.
- Li Y, Zhang X, Wang C, Hu Y, Niu X, Pei D, et al. Characterization by phenotypic and genotypic methods of metallo-β‑lactamase‑producing Pseudomonas aeruginosa isolated from patients with cystic fibrosis. Mol Med Rep. 2015; 11(1): 494-98.
- Shahcheraghi F, Nikbin VS, Feizabadi MM. Identification and genetic characterization of metallo-beta-lactamase-producing strains of Pseudomonas aeruginosa in Tehran, Iran. New Microbiol. 2010; 33(3): 243-48.
- Othman HB, Halim RMA, Abdul-Wahab HEE-a, Atta HA, Shaaban O. Pseudomonas aeruginosa-modified Hodge test (PAE-MHT) and ChromID Carba agar for detection of carbapenemase producing Pseudomonas aeruginosa recovered from clinical specimens. Open Access Macedonian Journal of Medical Sciences 2018; 6(12): 2283-290.
- Beig M, Taheri M, Arabestani MR. Frequency of metallo-β-lactamases and carbapenemase enzymes in clinical isolates of pseudomonas aeruginosa. 2020; 27(1): 21-9.
- Pena A, Donato A, Alves A, Leitao R, Cardoso O. Detection of Pseudomonas aeruginosa producing metallo-β-lactamase VIM-2 in a central hospital from Portugal. Euro J Clinic Microbiol Infect Dis. 2008; 27(12): 1269-271.
- Cheng X, Wang P, Wang Y, Zhang H, Tao C, Yang W, et al. Identification and distribution of the clinical isolates of imipenem-resistant Pseudomonas aeruginosa carrying metallo-β-lactamase and/or class 1 integron genes. J Huazhong Univ Sci Technol. 2008; 28(3): 235-38.
- Alkhudhairy MK, Al-Shammari MMM. Prevalence of metallo-β-lactamase–producing Pseudomonas aeruginosa isolated from diabetic foot infections in Iraq. New Microb New Infect. 2020; 35: 100661.
- Rahimzadeh Torabi L, Doudi M, Golshani Z. The frequency of blaIMP and blaVIM carbapenemase genes in clinical isolates of Pseudomonas aeruginosa in Isfahan medical centers. Med J Mashhad Univ Med Sci. 2016; 59(3): 139-47.
- Khosravi AD, Mihani F. Detection of metallo-β-lactamase–producing Pseudomonas aeruginosa strains isolated from burn patients in Ahwaz, Iran. Diagnos Microbiol Infect Dis. 2008; 60(1): 125-28.
Type of Study: Research |
Subject:
Bactriology
Received: 2020/12/22 | Accepted: 2021/08/22 | Published: 2021/08/1
Received: 2020/12/22 | Accepted: 2021/08/22 | Published: 2021/08/1
| Rights and permissions | |
 |
This work is licensed under a Creative Commons Attribution-NonCommercial 4.0 International License. |